Abstract
ERBB2 abnormalities frequently occur and serve as rationale therapeutic targets in cancer. In this study, clinical and next-generation sequencing data from 14,956 patients across more than 20 tumor types were collected. A total of 406 (2.7%) patients were identified with ERBB2 amplifications, and 303 (2.0%) patients with pathogenic somatic ERBB2 mutations. ERBB2 amplifications fell most frequently in breast (15.9%) and stomach (8.3%) cancers. Somatic ERBB2 SNVs/indels occurred most common in bladder/urinary tract (7.3%) and intestine (6.1%) cancers. The top mutated ERBB2 SNVs/indels were p.Y772_A775dup (25.5%) and p.S310F/Y (19.9%). Significantly higher rates of ERBB2 SNV/indels were found in women compared to men (2.8% vs. 1.5%, p < 0.0001). CDK12 was the most common co-amplification gene with ERBB2 in cancers with a high frequency of ERBB2 amplifications. Patients with ERBB2 amplifications or mutations had higher TMB compared with patients with non-ERBB2 alterations. The study provided the landscape of ERBB2 alterations across a variety of solid tumors that may benefit from anti-HER2 agents.
Introduction
ERBB2 gene, also known as HER2, is a well-known proto-oncogene encoding a receptor tyrosine kinase (RTK), which consists of an impaired extracellular ligand-binding structure containing two receptor-L domains and a furin-like cysteine-rich region, a single hydrophobic transmembrane domain and an intracellular tyrosine kinase domain (1–3). Alterations of ERBB2 including gene amplification, protein overexpression, and missense mutations have been reported in multiple solid cancers, especially in breast cancer (4), stomach cancer (5), and non-small cell lung cancer (NSCLC) (6). These aberrant activations of ERBB2, independent of ligand-receptor stimulation, promote tumorigenesis, tumor growth, and progression (7).
Since the first monoclonal antibody targeting the extracellular domain of HER2, Trastuzumab, got approved by the US Food and Drug Administration (FDA) for the treatment in HER2-positive breast cancer in 1998, a number of anti-HER2 agents have been developed and modified for the treatment of patients harboring ERBB2 amplifications, including monoclonal antibodies, small molecular drugs, and antibody-drug conjugates (ADCs). Trastuzumab in combination with chemotherapy has been approved for the first-line treatment of patients with HER2-positive metastatic gastric cancer since 2009 (8). The tyrosine kinase inhibitor lapatinib can bind to the ATP-binding pocket reversibly and suppress the RAS/RAF/MEK/ERK and PI3K-AKT signaling in HER2-positive solid cancers (9, 10). In recent years, the bispecific antibodies that covalently combine cytotoxic agents and monoclonal antibodies, i.e., antibody-drug conjugates (ADCs), have emerged as a novel class of targeted anti-cancer drug. Several ADCs derived from anti-HER2 mAbs have exhibited excellent clinical efficacy and have been approved for patients with solid cancers with ERBB2 amplification (11–15). On the other side, although no targeted drugs are approved for ERBB2-mutated cancers by far, several clinical trials have exhibited promising results in lung cancer patients carrying ERBB2 mutations. In a single-arm, open-label, phase II study, Poziotinib, an irreversible pan-HER inhibitor, showed hopeful antitumor activity with an ORR of 27% in patients with ERBB2 exon 20 mutant NSCLC including patients who had previously received platinum-based chemotherapy. The median progression-free survival (mPFS) was 5.5 months (95% CI, 4.0–7.0) and the median overall survival (mOS) was 15 months (95% CI, 9.0 to not estimable)(16). Based on the encouraging clinical efficacy of Poziotinib, FDA granted it fast-track designation. Results from a phase II basket trial displayed that Ado-trastuzumab emtansine provided a confirmed partial response rate of 44% (95% CI, 22%–69%) in 18 patients with advanced HER2-mutant lung adenocarcinomas and mPFS was 5 months (95% CI, 3–9 months). Responses were seen in patients with ERBB2 exon 20 insertions and point mutations in the kinase, transmembrane, and extracellular domains (17). Most recently, a multicenter, international, phase 2 study revealed that Trastuzumab deruxtecan showed durable antitumor activity with ORR of 55% in 91 patients with previously treated HER2-mutant NSCLC. mPFS was 8.2 months (95% CI, 6.0–11.9), and mOS was 17.8 months (95% CI, 13.8–22.1) (18). Therefore, the National Comprehensive Cancer Network (NCCN) guidelines for non-small cell lung cancer (NSCLC) recommend that testing should be conducted as part of broad molecular profiling to detect ERBB2 mutations in newly diagnosed patients and Ado-trastuzumab emtansine and Trastuzumab deruxtecan are available targeted agents in ERBB2-mutated NSCLC.
In the present study, we aim to explore the molecular landscape of ERBB2 alterations in solid tumors by next-generation sequencing (NGS), which will provide additional evidence for the panorama of these therapeutic targets in the Chinese cancer population.
Materials and Methods
Samples
Formalin-fixed paraffin-embedded (FFPE) samples from cancer patients who underwent NGS between January 2017 and June 2020 were screened for analysis. The clinicopathological information from the patients and their sequencing data were collected.
This study was conducted under the approval of the ethics committees of the hospitals and informed consent was obtained from patients.
NGS Detection
The methods for genome profiling have been described in the previous publication (19). Briefly, genomic profiling of DNA was performed through NGS with a >150-gene panel on Illumina Nextseq 500 to >500X coverage in 3DMed Clinical Laboratory Inc., a laboratory certified by both College of American Pathologists (CAP) and Clinical Laboratory Improvement Amendments (CLIA). Both somatic and germline alterations were analyzed, including single nucleotide variants (SNVs), insertion and deletions (indels), and copy number variants (CNVs). All SNVs and indels in the coding region of targeted genes, including missense, silent, stop gain, stop loss, in-frame, and frameshift mutations were considered. The CNVs of tumor tissues were calculated by BIC-seq2 (20). In addition, the TMB was defined as the number of nonsynonymous somatic SNVs and indels in examined coding regions, with driver mutations excluded. A microsatellite instability (MSI) score was defined as the percentage of unstable loci. Any sample with an MSI score of ≥0.4 was classified as MSI-H, and otherwise MSS. The identification of pathogenic levels of variants was obtained based on the published reports and the recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMP-AMP).
Statistical Analysis
Categorical variables were described as number and proportions. Categorical relationships were examined by using Pearson’s chi-square test with the Yates continuity correction when applicable and p value <0.05 was considered statistically significant. The SPSS22.0 software (SPSS, Inc., Chicago, IL, United States) was carried out for statistical analysis.
Results
A total of 14,956 cancer patients who have received tissue NGS with a panel of more than 150 cancer-related genes were included for analysis. The median age was 59 (IQR range, 10–94), and 60% were men. The patients carried more than 20 types of solid tumor, including lung cancer (n = 4,624, 30.9%), liver cancer (n = 2,021, 13.5%), colorectal cancer (n = 1,581, 10.6%), stomach cancer (n = 998, 6.7%), biliary tract cancer (n = 840, 5.6%), etc. Among 12,450 patients who were evaluable for MSI status, 258 (1.7%) presented with MSI-H (Table 1).
TABLE 1
| Characteristics | All patients (N = 14,956) |
|---|---|
| Age, median (IQR range) | 59 (10–94) |
| Sex, n (%) | |
| Male | 8,980 (60.0) |
| Female | 5,976 (40.0) |
| MSI status, n (%) | |
| MSI-H | 258 (1.7) |
| MSS | 12,192 (81.5) |
| N/A | 2,506 (16.8) |
| Tumor types, n (%) | |
| Lung cancer | 4,624 (30.9) |
| Liver cancer | 2021 (13.5) |
| Colorectal cancer | 1,581 (10.6) |
| Stomach cancer | 998 (6.7) |
| Biliary tract cancer | 840 (5.6) |
| Kidney cancer | 828 (5.5) |
| Pancreas cancer | 778 (5.2) |
| Breast cancer | 503 (3.4) |
| Sarcoma | 437 (2.9) |
| Ovary cancer | 394 (2.6) |
| Head and Neck Cancer | 340 (2.3) |
| Others | 306 (2) |
| Cervical cancer | 225 (1.5) |
| Esophagus cancer | 195 (1.3) |
| Intestine cancer | 179 (1.2) |
| Bladder/urinary tract cancer | 177 (1.2) |
| Endometrium cancer | 152 (1) |
| Prostate cancer | 131 (0.9) |
| Neuroendocrine carcinoma | 128 (0.9) |
| Gastrointestinal stromal tumors | 119 (0.8) |
Clinicopathological information of the cancer patients.
Overall, 303 (2.1%) patients were identified with 321 pathogenic or very likely pathogenic somatic ERBB2 SNVs/indels, including 213 missense mutations and 108 in-frame insertions or deletions, identifying 46 unique SNV/indel mutations. In addition, germline ERBB2 alterations were found in 157 (1.0%) patients. Amongst all, 40 patients harbored at least two somatic ERBB2 mutations, including 25 with concomitant ERBB2 amplifications and mutations. Somatic ERBB2 SNVs/indels occurred most common in bladder/urinary tract cancer (13/177, 7.3%), intestine cancer (11/179, 6.1%), stomach cancer (41/998, 4.1%), endometrium cancer (5/152, 3.3%), and lung cancer (125/4,624, 2.7%). Of note, SNVs were the majority of variants in almost all the cancers, except for lung cancer in which indels were the main variants. On the other side, the mutational frequency of ERBB2 was the lowest in kidney cancer, sarcoma, liver cancer, and ovary cancer (Figure 1A). Across all cancers, ERBB2 mutations occurred most frequently in the tyrosine kinase domain (62.3%) which included mutations in exon 19 (9.7%), exon 20 (38.3%), exon 21 (11.8%), and others (2.5%), followed by a furin-like cysteine-rich region of the extracellular domain (20.6%) and transmembrane domain (17.1%, Figure 1B). Of note, ERBB2 exon 20 insertions were detected in 93 (0.6%) patients, accounting for 29.0% of all detected ERBB2 SNV/indels, including 86 with lung cancer, two with pancreas cancer, one with breast cancer, etc. The top mutated ERBB2 SNVs/indels were ERBB2 p.Y772_A775dup (82/321, 25.5%), a type of in-frame insertion within the exon 20 of the tyrosine kinase domain, and p.S310F/Y (64/321, 19.9%) in furin-like cysteine-rich region, followed by p.R678Q (43/321, 13.4%) in the transmembrane domain, and p.V842I (26/321, 8.1%) in the tyrosine kinase domain, etc. (Figure 1C; Table 2). In addition, the form and frequency of each HER2 variant was observed for different tumor sites (Supplementary Table S1).
FIGURE 1
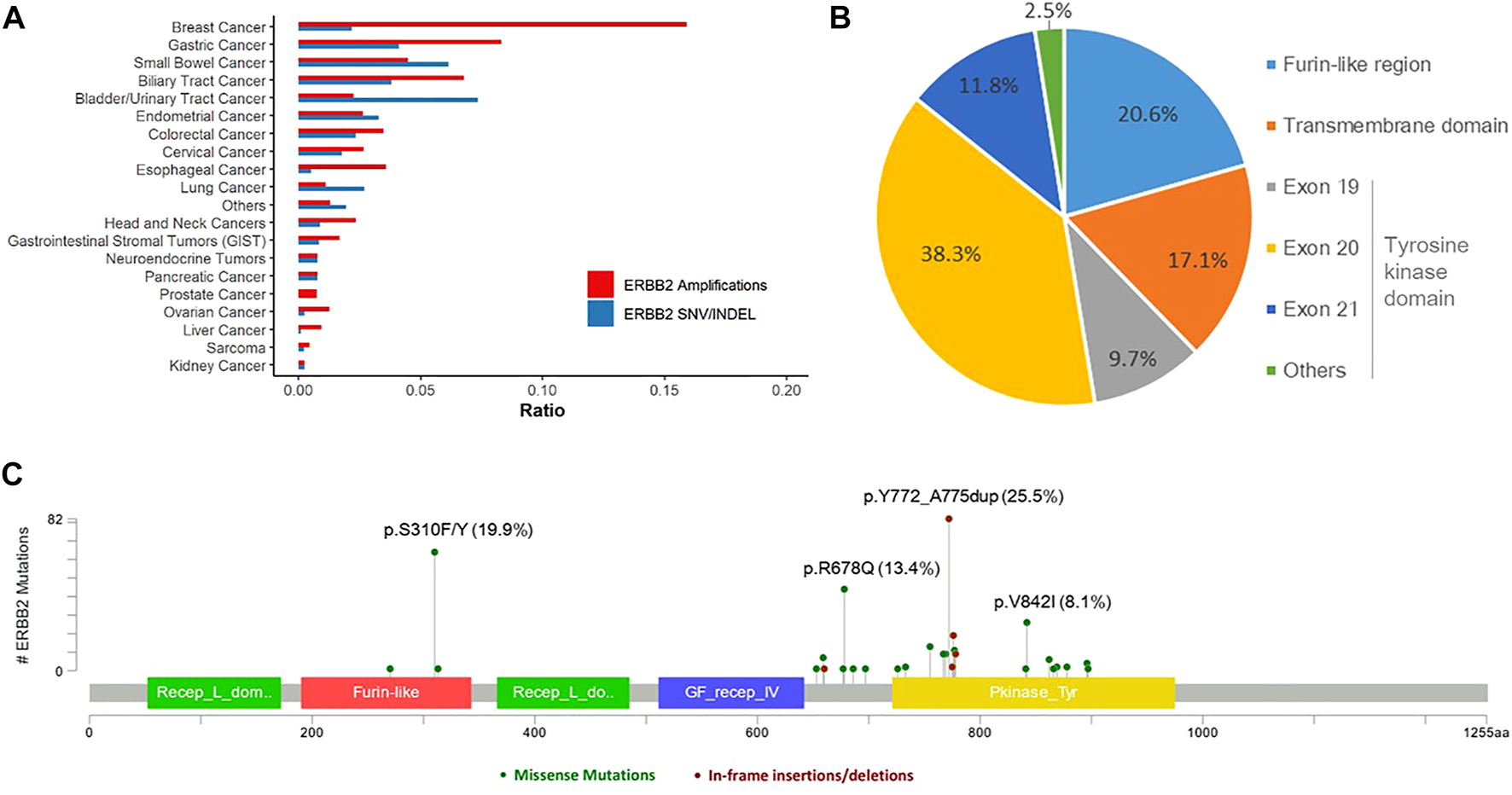
Landscape of ERBB2 alterations. (A) Landscape of ERBB2 amplifications and ERBB2 mutations across different tumor types. (B) Prevalence of the pathogenic or very likely pathogenic somatic mutations of ERBB2 in furin-like region, transmembrane domain, and tyrosine kinase domain. (C)ERBB2 somatic mutation map.
TABLE 2
| SNV/indels | Counts | Percent (%) |
|---|---|---|
| p.Y772_A775dup | 82 | 25.5 |
| p.S310F | 47 | 14.6 |
| p.R678Q | 43 | 13.4 |
| Others | 27 | 8.4 |
| p.V842I | 26 | 8.1 |
| p.S310Y | 17 | 5.3 |
| p.V777L | 11 | 3.4 |
| p.G776delinsVC | 10 | 3.1 |
| p.L755S | 9 | 2.8 |
| p.G778_P780dup | 8 | 2.5 |
| p.I767M | 8 | 2.5 |
| p.D769Y | 7 | 2.2 |
| p.V659E | 7 | 2.2 |
| p.T862A | 5 | 1.6 |
| p.G776V | 3 | 0.9 |
| p.L755P | 3 | 0.9 |
| p.D769H | 2 | 0.6 |
| p.H878Y | 2 | 0.6 |
Prevalence of the pathogenic or very likely pathogenic somatic mutations of ERBB2.
Among several cancers in which ERBB2 mutation occurred most common, we investigated the genetic variation spectrum. In bladder/urinary tract cancer, ARID1A (54%), TP53 (54%), and FGFR3 (38%) were the high-frequency mutated genes (Figure 2A), whereas TP53 (64%), ARID1A (36%), and CTNNB1 (36%) were common genes observed in intestine cancer (Figure 2B). Furthermore, TP53 (61%), ARID1A (37%), and LRP1B (32%) were the top three mutated genes in stomach cancer (Figure 2C), while in lung cancer, TP53 (46%), LRP1B (10%), and SPTA1 (10%) were the most frequently mutated genes (Figure 2D). We further explored whether ERBB2 mutation in different domains would lead to a difference in mutation frequency in the variation spectrum. Analysis indicated that CDKN2A, FAT1, MSH6, ARID1A, KEAP1, NF1, and SMAD4 mutation was more recurrent in patients with ERBB2 SNV/indels within the kinase domain compared with non-kinase domain in lung cancer (Figure 2E).
FIGURE 2
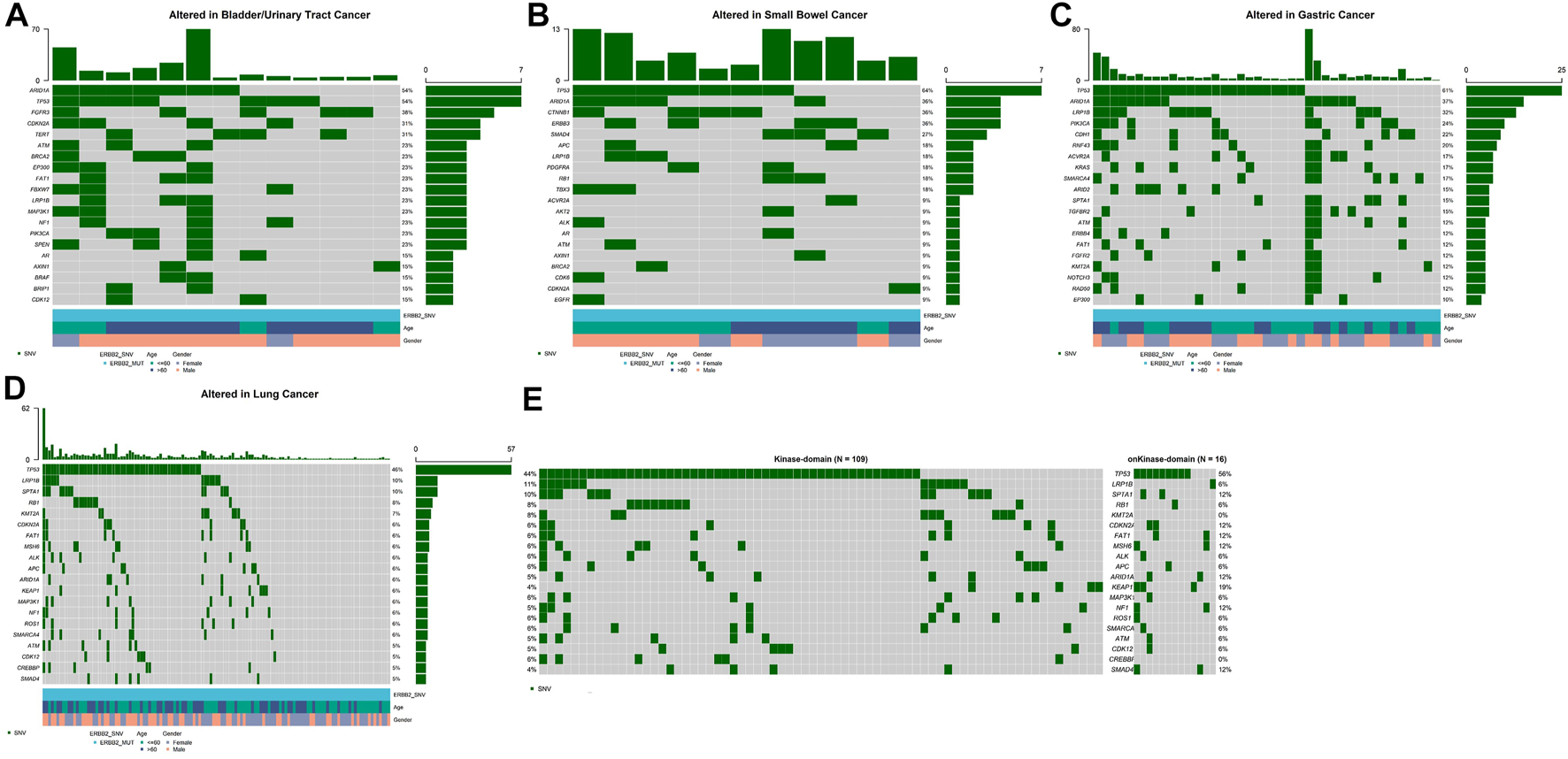
Top 20 high-frequency mutated genes in ERBB2-mutated patients of bladder/urinary tract cancer (A), intestine cancer (B), stomach cancer (C), lung cancer (D), kinase domain, and non-kinase domain in lung cancer (E).
Meanwhile 406 (2.7%) patients were identified with ERBB2 amplifications. ERBB2 amplifications fell most frequently in breast cancer (80/503, 15.9%), stomach cancer (83/998, 8.3%), biliary tract cancer (57/840, 6.8%), intestine cancer (8/179, 4.5%), and esophagus cancer (7/195, 3.6%, Figure 1A). Among ERBB2-amplification patients, except for TP53 which was the top most mutated gene, PIK3CA ranked second of high frequency mutated genes in breast cancer (Figure 3A) and mutation of PREX2 was recurrent in stomach cancer (Figure 3B). In biliary tract cancer, SMAD4 was the more frequent mutated gene (Figure 3C). KRAS showed a decreased mutation frequency in colorectal cancer patients with ERBB2 amplification (Figure 3D) compared with non-ERBB2 amplification (15% vs. 49%). On the other hand, CDK12 was the most common co-amplification gene with ERBB2 in breast cancer (59/80, 73.8%, Figure 3E), stomach cancer (37/83, 44.6%, Figure 3F), biliary tract cancer (33/57, 57.9%) (Figure 3G) and colorectal cancer (33/55, 60%, Figure 3H).
FIGURE 3
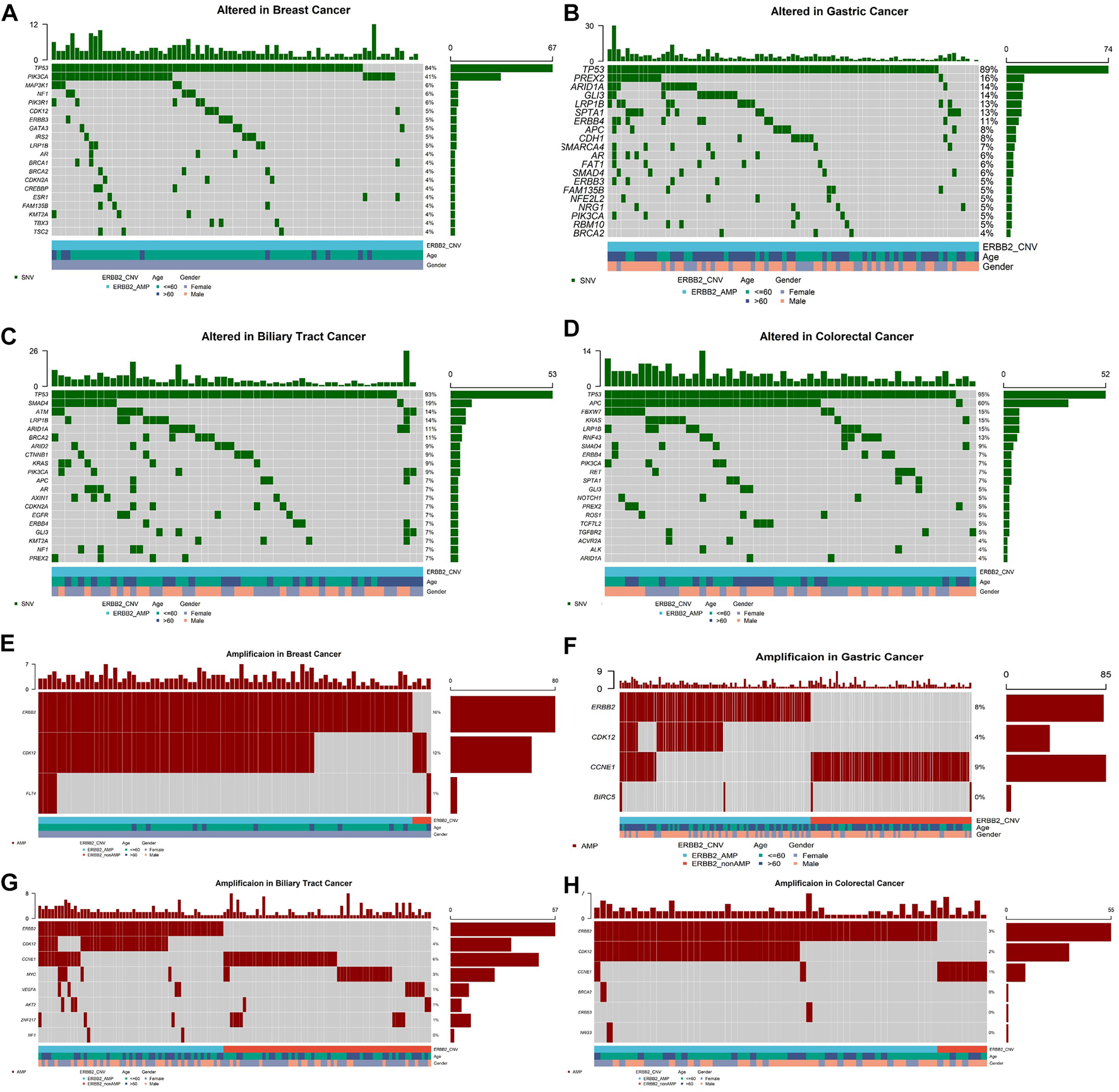
Top 20 high-frequency mutated genes in ERBB2-amplification patients of breast cancer (A), stomach cancer (B), biliary tract cancer (C), colorectal cancer (D), and co-amplification genes accompanying ERBB2 amplification in breast cancer (E), stomach cancer (F), biliary tract cancer (G), colorectal cancer (H).
No association was observed between the incidence of ERBB2 mutations and age (>60 vs. ≤60, p = 0.95 for SNVs/indels and p = 0.21 for amplifications). Significantly higher rates of ERBB2 alterations were found in female patients (female vs. male, mutations, 2.8% vs. 1.5%, p < 0.0001; amplifications, 3.7% vs. 2.1%, p < 0.0001). Given the higher incidence of ERBB2 mutations in breast cancer and gynecologic cancer, the analysis was re-conducted with these cancers being excluded, which showed a consistent higher rate in female patients compared to male patients (3.1% vs. 1.5%, p < 0.0001 for SNVs/indels and 2.7% vs. 2.1%, p = 0.037 for amplifications).
We further analyzed the association between ERBB2 variant and TMB. Patients without ERBB2 alterations had significantly lower TMB compared with ERBB2 mutation (p = 1.6e−0.6) (Figure 4A) and ERBB2 amplification (p = 0.0024) (Figure 4B) respectively in overall population. In a more detailed analysis, we found patients with ERBB2 mutation had significantly higher TMB compared with non-ERBB2 mutation in bladder/urinary tract cancer (p = 0.0045, Figure 4C), stomach cancer (p = 0.0045, Figure 4D), and colorectal cancer (p = 0.0021, Figure 4E) respectively. In contrast, patients with ERBB2 mutation had lower TMB compared with non-ERBB2 mutation in lung cancer (p = 0.048, Figure 4F). However, when we analyzed the association between ERBB2 amplification and TMB in several cancers, no significant difference was found (Supplementary Figure S1). We speculated that the small sample size in every cancer analyzed may be one reason. On the other hand, ERBB2 amplification detected by NGS was not totally equal to the status of HER2-positive detected by IHC in breast and stomach cancer. We also explored the relationship between MSI-H and ERBB2 mutation. Patients accompanying with MSI-H were mostly in the ERBB2 mutation group (Supplementary Figure S2).
FIGURE 4
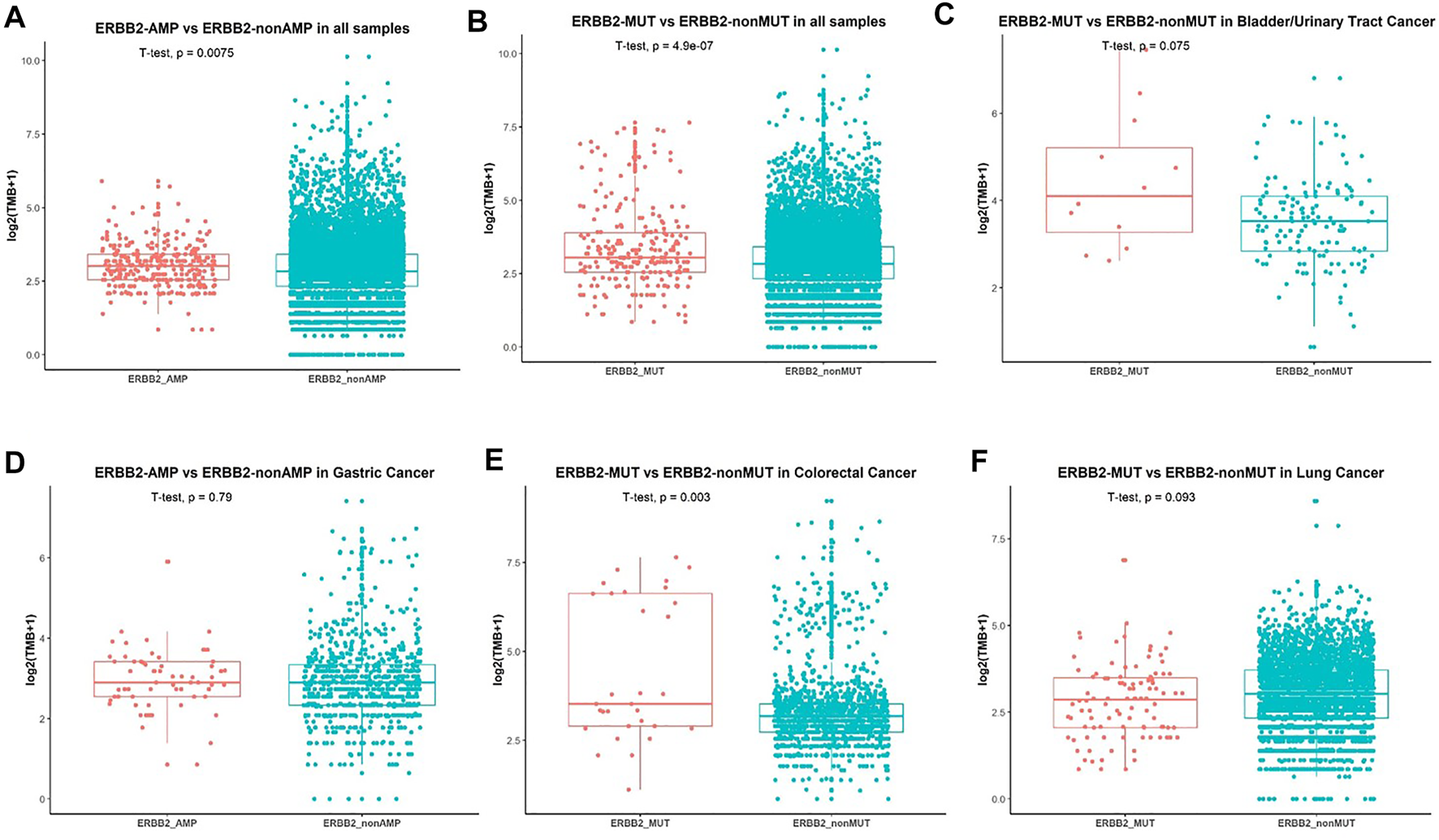
Association between ERBB2 alterations and TMB. (A) Comparison of TMB in patients with ERBB2-amplification and non-ERBB2-amplification. (B) Comparison of TMB in all patients with ERBB2-mutation and non-ERBB2-mutation and in bladder/urinary tract cancer (C), stomach cancer (D), colorectal cancer (E), lung cancer (F).
Discussion
In the present cross-sectional study, we explored the mutational landscape of ERBB2 in 14,956 pan-cancer patients. Our study showed a varying pattern of ERBB2 mutations across different tumor types. Overall, the distribution and incidence of ERBB2 amplifications and mutations in our Chinese cohort were mostly consistent with that in the Caucasian population derived from databases including the Catalogue of Somatic Mutations (COSMIC) and The Cancer Genome Atlas (TCGA) (21–24).
The identification of ERBB2 amplifications across tumors will provide evidence for the utilization of targeted agents such as trastuzumab, pertuzumab, T-DM1, lapatinib, neratinib, and the trastuzumab biosimilars. A study about HER2-positive breast cancer revealed the relationship between HER2 activity and the pro-trastuzumab tumor immune microenvironment. Gene expression profiling and immunohistochemistry analysis of 53 HER2-positive breast cancer patients indicated that trastuzumab-sensitive tumors expressed significantly higher levels of chemokines involved in immune cell recruitment, with higher infiltration of T cells and monocytes (25). The results were further supported by the recent results from a phase II trial, in which 37 treatment-naïve patients with HER2-positive metastatic oesophagogastric cancer were treated with a combination of pembrolizumab with trastuzumab and chemotherapy, providing numerically higher efficacy (objective response rate, 91%; median overall survival, 27.3 months) compared to the existing first-line standard (chemotherapy plus trastuzumab, objective response rate, 47%; median overall survival, 13.8 months) (8, 26). In this study, we found that patients with ERBB2 amplification had higher TMB than non-ERBB2 amplification (p = 0.0024), which provided new evidence for the application of immunotherapy in ERBB2 amplification cancers.
Besides, ERBB2 exon 20 insertions were found in 0.6% of the entire population and 1.9% of lung cancer patients. The most frequently appearing subtypes were Y772_A775dup (82/93, 88.2%) and p.G778_P780dup (8/93, 8.6%). ERBB2 exon 20 insertions were reported as analogous to EGFR exon 20 insertions and associated with primary resistance to currently approved tyrosine kinase inhibitors because of steric hindrance in the drug-binding pocket and a poor response to immunotherapies in lung adenocarcinoma (27, 28) _ENREF_24. In further analysis, ERBB2 exon 20-mutated tumors exhibited overexpression of RIPK1 and STK11IP and a decrease of cytotoxic natural killer cells (28). We also found lung cancer patients with ERBB2-mutant have lower TMB than non-ERBB2 mutant patients (p = 0.048). Taken together, these data suggest that ERBB2-mutant lung cancer patients may not benefit from immunotherapy. Whether immunotherapy can be used in other cancers carrying ERBB2 mutations/amplifications need further exploration.
In conclusion, we revealed a pan-cancer molecular landscape of ERBB2 amplification and mutations, and patients with ERBB2 alterations had higher TMB.
Statements
Data availability statement
The original presentations in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author. The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Nanjing Drum Tower Hospital. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author contributions
HW: Conceptualization, methodology, investigation, formal analysis, writing-original draft; JM: Conceptualization, methodology, investigation, formal analysis, writing—original draft; YW: Methodology, investigation, and formal analysis; XX: Data curation, formal analysis, visualization, writing—review and editing; YC: Data Curation, formal analysis, visualization, and software; MH: Formal analysis, visualization, writing—review and editing; SC: Data curation and formal analysis; ZZ: Data curation and formal analysis; YZ: Data curation and formal analysis; XZ: Conceptualization, funding acquisition, resources, supervision, writing—review and editing.
Funding
The work was supported by the Jiangsu Province Government Foundation (2016-WSW-067).
Conflict of interest
XX, YC, MH, SC, ZZ, YZ, and CC were employed by 3D Medicines Inc.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.por-journal.com/articles/10.3389/pore.2022.1610360/full#supplementary-material
References
1.
Yarden Y Sliwkowski MX . Untangling the ErbB Signalling Network. Nat Rev Mol Cel Biol (2001) 2(2):127–37. 10.1038/35052073
2.
Prenzel N Fischer OM Streit S Hart S Ullrich A . The Epidermal Growth Factor Receptor Family as a central Element for Cellular Signal Transduction and Diversification. Endocr Relat Cancer (2001) 8(1):11–31. 10.1677/erc.0.0080011
3.
Roskoski R . The ErbB/HER Family of Protein-Tyrosine Kinases and Cancer. Pharmacol Res (2014) 79:34–74. 10.1016/j.phrs.2013.11.002
4.
Slamon DJ Clark GM Wong SG Levin WJ Ullrich A McGuire WL . Human Breast Cancer: Correlation of Relapse and Survival with Amplification of the HER-2/Neu Oncogene. Science (1987) 235(4785):177–82. 10.1126/science.3798106
5.
Wang YK Gao CF Yun T Chen Z Zhang XW Lv XX et al Assessment of ERBB2 and EGFR Gene Amplification and Protein Expression in Gastric Carcinoma by Immunohistochemistry and Fluorescence In Situ Hybridization. Mol Cytogenet (2011) 4(1):14. 10.1186/1755-8166-4-14
6.
Wei XW Gao X Zhang XC Yang JJ Chen ZH Wu YL et al Mutational Landscape and Characteristics of ERBB2 in Non-small Cell Lung Cancer. Thorac Cancer (2020) 11(6):1512–21. 10.1111/1759-7714.13419
7.
Emde A Köstler WJ Yarden Y . Association of Radiotherapy and Oncology of the Mediterranean arEa. Therapeutic Strategies and Mechanisms of Tumorigenesis of HER2-Overexpressing Breast Cancer. Crit Rev Oncol Hematol (2012) 84(Suppl. 1):e49–e57. 10.1016/j.critrevonc.2010.09.002
8.
Bang Y-J Van Cutsem E Feyereislova A Chung HC Shen L Sawaki A et al Trastuzumab in Combination with Chemotherapy versus Chemotherapy Alone for Treatment of HER2-Positive Advanced Gastric or Gastro-Oesophageal junction Cancer (ToGA): A Phase 3, Open-Label, Randomised Controlled Trial. Lancet (2010) 376(9742):687–97. 10.1016/S0140-6736(10)61121-X
9.
Konecny GE Pegram MD Venkatesan N Finn R Yang G Rahmeh M et al Activity of the Dual Kinase Inhibitor Lapatinib (GW572016) against HER-2-Overexpressing and Trastuzumab-Treated Breast Cancer Cells. Cancer Res (2006) 66(3):1630–9. 10.1158/0008-5472.CAN-05-1182
10.
O'Brien NA Browne BC Chow L Wang Y Ginther C Arboleda J et al Activated Phosphoinositide 3-kinase/AKT Signaling Confers Resistance to Trastuzumab but Not Lapatinib. Mol Cancer Ther (2010) 9(6):1489–502. 10.1158/1535-7163.MCT-09-1171
11.
Verma S Miles D Gianni L Krop IE Welslau M Baselga J et al Trastuzumab Emtansine for HER2-Positive Advanced Breast Cancer. N Engl J Med (2012) 367(19):1783–91. 10.1056/NEJMoa1209124
12.
Krop IE Kim S-B González-Martín A LoRusso PM Ferrero J-M Smitt M et al Trastuzumab Emtansine versus Treatment of Physician's Choice for Pretreated HER2-Positive Advanced Breast Cancer (TH3RESA): A Randomised, Open-Label, Phase 3 Trial. Lancet Oncol (2014) 15(7):689–99. 10.1016/S1470-2045(14)70178-0
13.
Perez EA Barrios C Eiermann W Toi M Im Y-H Conte P et al Trastuzumab Emtansine with or without Pertuzumab versus Trastuzumab Plus Taxane for Human Epidermal Growth Factor Receptor 2-Positive, Advanced Breast Cancer: Primary Results from the Phase III MARIANNE Study. J Clin Oncol (2017) 35(2):141–8. 10.1200/JCO.2016.67.4887
14.
Modi S . Trastuzumab Deruxtecan in Previously Treated HER2-Positive Metastatic Breast Cancer: Plain Language Summary of the DESTINY-Breast01 Study. Future Oncol (2021) 17(26):3415–23. 10.2217/fon-2021-0427
15.
Shitara K Bang Y-J Iwasa S Sugimoto N Ryu M-H Sakai D et al Trastuzumab Deruxtecan in Previously Treated HER2-Positive Gastric Cancer. N Engl J Med (2020) 382(25):2419–30. 10.1056/NEJMoa2004413
16.
Elamin YY Robichaux JP Carter BW Altan M Gibbons DL Fossella FV et al Poziotinib for Patients with HER2 Exon 20 Mutant Non-small-cell Lung Cancer: Results from a Phase II Trial. J Clin Oncol (2022) 40:702–9. 10.1200/JCO.21.01113
17.
Li BT Shen R Buonocore D Olah ZT Ni A Ginsberg MS et al Ado-Trastuzumab Emtansine for Patients with HER2-Mutant Lung Cancers: Results from a Phase II Basket Trial. J Clin Oncol (2018) 36(24):2532–7. 10.1200/JCO.2018.77.9777
18.
Li BT Smit EF Goto Y Nakagawa K Udagawa H Mazières J et al Trastuzumab Deruxtecan in HER2-Mutant Non-small-cell Lung Cancer. N Engl J Med (2022) 386:241–51. 10.1056/NEJMoa2112431
19.
Su D Zhang D Chen K Lu J Wu J Cao X et al High Performance of Targeted Next Generation Sequencing on Variance Detection in Clinical Tumor Specimens in Comparison with Current Conventional Methods. J Exp Clin Cancer Res (2017) 36(1):121. 10.1186/s13046-017-0591-4
20.
Xi R Lee S Xia Y Kim T-M Park PJ . Copy Number Analysis of Whole-Genome Data Using BIC-Seq2 and its Application to Detection of Cancer Susceptibility Variants. Nucleic Acids Res (2016) 44(13):6274–86. 10.1093/nar/gkw491
21.
Gao J Aksoy BA Dogrusoz U Dresdner G Gross B Sumer SO et al Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci Signal (2013) 6(269):pl1. 10.1126/scisignal.2004088
22.
Cerami E Gao J Dogrusoz U Gross BE Sumer SO Aksoy BA et al The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov (2012) 2(5):401–4. 10.1158/2159-8290.CD-12-0095
23.
Robertson AG Kim J Al-Ahmadie H Bellmunt J Guo G Cherniack AD et al Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell (2018) 174(4):1033. 10.1016/j.cell.2018.07.036
24.
Robichaux JP Elamin YY Vijayan RSK Nilsson MB Hu L He J et al Pan-Cancer Landscape and Analysis of ERBB2 Mutations Identifies Poziotinib as a Clinically Active Inhibitor and Enhancer of T-DM1 Activity. Cancer Cell (2019) 36(4):444–57. 10.1016/j.ccell.2019.09.001
25.
Triulzi T Forte L Regondi V Di Modica M Ghirelli C Carcangiu ML et al HER2 Signaling Regulates the Tumor Immune Microenvironment and Trastuzumab Efficacy. Oncoimmunology (2019) 8(1):e1512942. 10.1080/2162402X.2018.1512942
26.
Janjigian YY Maron SB Chatila WK Millang B Chavan SS Alterman C et al First-line Pembrolizumab and Trastuzumab in HER2-Positive Oesophageal, Gastric, or Gastro-Oesophageal junction Cancer: An Open-Label, Single-Arm, Phase 2 Trial. Lancet Oncol (2020) 21(6):821–31. 10.1016/S1470-2045(20)30169-8
27.
Robichaux JP Elamin YY Tan Z Carter BW Zhang S Liu S et al Mechanisms and Clinical Activity of an EGFR and HER2 Exon 20-selective Kinase Inhibitor in Non-small Cell Lung Cancer. Nat Med (2018) 24(5):638–46. 10.1038/s41591-018-0007-9
28.
Kirchner M Kluck K Brandt R Volckmar A-L Penzel R Kazdal D et al The Immune Microenvironment in EGFR- and ERBB2-Mutated Lung Adenocarcinoma. ESMO Open (2021) 6(5):100253. 10.1016/j.esmoop.2021.100253
Summary
Keywords
next-generation sequencing, TMB, ERBB2 , solid tumors, anti-HER2 agents
Citation
Wang H, Miao J, Wen Y, Xia X, Chen Y, Huang M, Chen S, Zhao Z, Zhang Y, Chen C and Zhu X (2022) Molecular Landscape of ERBB2 Alterations in 14,956 Solid Tumors. Pathol. Oncol. Res. 28:1610360. doi: 10.3389/pore.2022.1610360
Received
16 February 2022
Accepted
23 June 2022
Published
13 July 2022
Volume
28 - 2022
Edited by
József Tímár, Semmelweis University, Hungary
Updates
Copyright
© 2022 Wang, Miao, Wen, Xia, Chen, Huang, Chen, Zhao, Zhang, Chen and Zhu.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinhua Zhu, drzhuxh@163.com
†These authors have contributed equally to this work and share first authorship
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.