Abstract
Aims: Metastatic cervical carcinoma is hard to cure using traditional treatment and new therapeutic approaches are needed. However, the process of clonal evolution and the molecular alterations that contribute to tumor progression from primary to metastatic carcinoma remain unclear. It is currently difficult to distinguish between the primary pulmonary squamous cell carcinoma (PPSCC) and metastatic cervical squamous cell carcinoma (CSCC).
Methods: Paired primary CSCC and lung/lymph nodes metastatic lesions from eight patients were analyzed by whole-exome sequencing (WES). WES data of matched specimens and normal samples were aligned to the human reference genome and analyzed to identify somatic mutations in primary and metastatic lesions.
Results: A total of 1,254 somatic variants were identified. All the primary lesions and metastatic lesions shared mutations, the percentage of shared mutations between primary lesions and corresponding metastatic lesions varied significantly, ranging from 6% to 70%. In other words, all the metastatic lesions are clonally related to primary lesions, confirming WES could prove they are metastatic from the cervix but not PPSCC. We tried to apply a gene panel to help distinguish PPSCC and metastatic CSCC but failed because the mutations were widely distributed in CSCC. Interestingly, lymph nodes metastasis (LNM) harbored fewer cancer driver mutations than primary CSCC specimens with a significant difference. Besides this, there was no significant difference in somatic mutations and copy number variation (CNV) between primary and metastatic CSCC.
Conclusion: Our data demonstrate that WES is an additional helpful tool in distinguishing PPSCC and metastatic CSCC, especially for certain difficult cases.
Introduction
Cervical carcinoma is the third most common gynecologic cancer and the fourth leading cause of cancer death worldwide [1]. Human papillomavirus is central to the development of cervical neoplasia and can be detected in more than 90 percent of cervical cancers [2]. The most common histologic types of cervical cancer are CSCC. The pelvic nodes and lung are the most frequent metastatic sites for CSCC [3]. For patients with cervical cancer, 4.16–7.7% of patients have lung metastasis [4].
One problem that exists in our routine work is that it is hard to distinguish between metastatic CSCC and primary lesion. If a solitary lung squamous cell carcinoma is found in a patient with CSCC history, the lesion could be either metastasis CSCC or a PPSCC. The differential diagnosis has a big difference in treatment and prognosis. Lung metastasis (LM) is formed through hematogenous metastasis of CSCC and originates from primary CSCC clones. At the same time, PPSCC is generally believed to originate from independent clones from primary CSCC. The treatment is also different for the two diseases; further chemotherapy is needed for LM, while PPSCC cases require surgery. Thus, distinguishing PPSCC from LM is important since the diagnosis can affect subsequent treatment strategies and survival evaluations. In clinical practice, we compare the morphology similarity of lung tumor and CSCC; if they are similar, we tend to believe they are metastatic lesions, and vice versa. However, the morphology is unreliable because the CSCC and PPSCC could be pretty similar. And the p16 positive expression can’t help differential diagnosis because the p16 staining could also be strongly positive in PPSCC cases [5–7]. HPV genotyping [7, 8] could only discriminate about half of these controversial cases because PPSCC could also have HPV infection status. Thus, in this study, we tried to apply WES and analyze if they are clonally related to help distinguish LM and PPSCC.
Metastatic cervical carcinoma is hard to cure using traditional treatment, so new therapeutic approaches are needed. Previous studies demonstrated the function of PTEN, PIK3CA, TP53, and KRAS in the pathogenesis of CSCC [9–12]. The genomic landscape was well established in 2017 by The Cancer Genome Atlas (TCGA) Research Network, which increased our understanding of CSCC [13]. They observed some novel mutations in CSCC, like SHKBP1, CASP8, HLA-A, and TGFBR2. Despite this advancement, a comprehensive molecular landscape depicting the genome alterations during the tumor progression from primary CSCC to metastatic diseases remains elusive. The molecular pathogenesis of metastatic CSCC and how primary CSCC progresses to metastatic CSCC remain largely unclear as genome-wide molecular genetic analysis of metastatic CSCC has not been reported. Revealing this could help us acquire a deep understanding of the genomic process of metastasis and may help us develop new targets on preventing metastasis and improving survival. This study performed WES in surgical specimens of eight pairs of primary and metastatic CSCC. And we analyzed somatic mutations including cancer driver mutations and CNV between primary and metastatic CSCC.
Materials and Methods
Tumor Specimen
All eight cases were selected from Cancer Hospital, Chinese Academy of Medical Sciences after approval by the Institutional Review Board. The sequence cohort was selected based on the availability of paired tumor primary and metastatic tissues and matched normal tissue controls. All cases received the total hysterectomy surgery. Two experienced pathologists confirmed the diagnosis (L. L. and Y. S.). CSCC1-CSCC3 had paired primary CSCCs and LM specimens, and CSCC4-CSCC8 had paired primary pelvic LNM samples. CSCC1 had two LM specimens, which were LM1 and LM2. The interval from LM1 to LM2 was 12 months for case 1. Finally, 25 FFPE samples, including the paired normal control samples from eight patients with CSCCs (CSCC1- CSCC8), were submitted to WES.
Somatic Mutations in Paired Primary and Metastatic CSCC
Tumor and normal DNA were extracted from tissue shavings of formalin-fixed and paraffin-embedded specimens using QIAamp DNA FFPE Tissue Kit (Qiagen, Valencia, CA, United States) according to the manufacturer’s instructions. DNA was quantified by Qubit (Life Technologies).
According to the manufacturer’s instructions, WES was performed on the genomic DNA obtained from tumor tissues and matched normal tissue using the Agilent Sure Select Human All Exome V5 Kit (Agilent Technologies, Santa Clara, CA, United States). Paired-end sequencing, resulting in 150 bases from each end of the fragments, was performed using the HiSeq X Ten. The paired-end sequencing raw data (FASTQ format) were aligned to the human reference genome (hg19) using Burrows-Wheeler aligner software (BWA, v0.7.15). The Genome Analysis Toolkit (GATK, v3.6), Picard (http://picard.sourceforge.net; v2.7.1), and Samtools (v1.3.1) were used for the basic processing and management of marking duplicates, including local realignments and score recalibration. Somatic mutations were detected by comparing tumor and matched normal sequencing data using MuTect1 and Strelka for single-nucleotide variants and InDels. Somatic mutations in the test samples were defined according to the following criteria: 1) the mutation was identified in four or more distinct reads and the mutation depth was greater than 15 reads, 2) the mutation was not present in any of the reads in the matched control sample or the other control samples analyzed in this study, and 3) the mutation was uncommon in dbSNPs (frequency < 1%). Only non-synonymous were reported. Putative driver mutations were determined based on previously listed cancer driver genes [14]. All candidate somatic mutations were validated by visual inspection using the Integrated Genome Viewer (IGV) [15].
Moreover, ANNOVAR was used for the functional annotation of each variant in the coding regions. PolyPhen-2 was utilized to predict the impact of an amino acid substitution on protein function and structure.
Detecting Copy Number Variation and Clonal Composition Using WES
Binned copy number and segmentation data of the tumor genome, compared to the matched normal tissue, were computed using the copy number calling pipeline of alignment data in the CNVkit package [16]. The visualization of log-ratio segments was done using matplotlib plotting library.
Phylogenetic Analysis
We used the “discrete‐characters Wagner parsimony” method in phylogeny inference package (phylip) version 3.698 [17] to generate phylogenetic trees. All phylogenetic trees were rooted using the control tissue DNA, which represents the ancestral state. Cladograms were plotted with the number of mutations per branch at each end. The trees were drawn using the DrawTree tool under the phylip package.
Patient and Public Involvement Statement
Patients were involved in the design and conduct of our research.
Statistical Analysis
Statistical analysis was done with Statistical Package for the Social Sciences (SPSS 22.0). In all experiments, comparisons between two groups were based on the Wilcoxon test. Mann-Whitney U was utilized to test for differences among groups. p-values of <0.05 were considered statistically significant.
Results
Clinicopathological Characteristics
The clinicopathological information of the eight CSCC patients was summarized in Table 1. The median age of these patients was 48 years old. The histopathological features in a representative case are illustrated in Supplementary Figure S1. Four patients had poorly differentiated carcinomas, three had moderately differentiated carcinomas, and one had well-differentiated carcinoma. International Federation of Gynecology and Obstetrics (FIGO) stage is IB1 (1/8), IB2 (5/8), and IIA (2/8), respectively. The average months from the primary tumor to the LM tumor was 37 months. All patients received neoadjuvant therapy after the surgery.
TABLE 1
| Case ID | Gender | Age of first-time diagnosis (yrs.) | Smoking status | FIGO stage | Histology | Differentiation | Present with distance metastases at first-time diagnosis |
|---|---|---|---|---|---|---|---|
| CSCC1 | Female | 45–50 | No | IIa2 | SCC | Poor | Y |
| CSCC2 | Female | 35–40 | No | Ib2 | SCC | Moderate | Y |
| CSCC3 | Female | 50–55 | No | Ib2 | SCC | Poor | Y |
| CSCC4 | Female | 45–50 | No | Ib2 | SCC | Moderate | N |
| CSCC5 | Female | 45–50 | No | Ib2 | SCC | Poor | N |
| CSCC6 | Female | 45–50 | No | Ib2 | SCC | Well | N |
| CSCC7 | Female | 50–55 | No | IIa1 | SCC | Poor | N |
| CSCC8 | Female | 45–50 | No | Ib1 | SCC | Moderate | N |
Clinicopathological characteristics of eight patients.
The Comparison Between Primary and Metastatic CSCC Carcinoma
As shown in Supplementary Table S1, the average target depth for the analysis was 182.3×, 98.9% of which was covered by more than ten reads. One thousand two hundred fifty-four non-synonymous somatic mutations were identified. The complete list of somatic variants was presented in Supplementary Table S2. From these, 91 canonical cancer-driver mutations were detected (Supplementary Table S3).
The mutation number of eight CSCC pairs is shown in Figure 1A. Interestingly, the LNM harbored fewer mutations than primary CSCC specimens, although the difference was insignificant (p = 0.08, Wilcoxon test). The LM specimens also had more mutations than the LNM specimens without significant difference (p = 0.07, Wilcoxon test). In the Venn diagram for the eight cases (Figure 1B), the paired primary lesions and LM shared 28%–70% of mutated genes, while the paired primary lesions and LNM shared 6–47% of mutated genes.
FIGURE 1
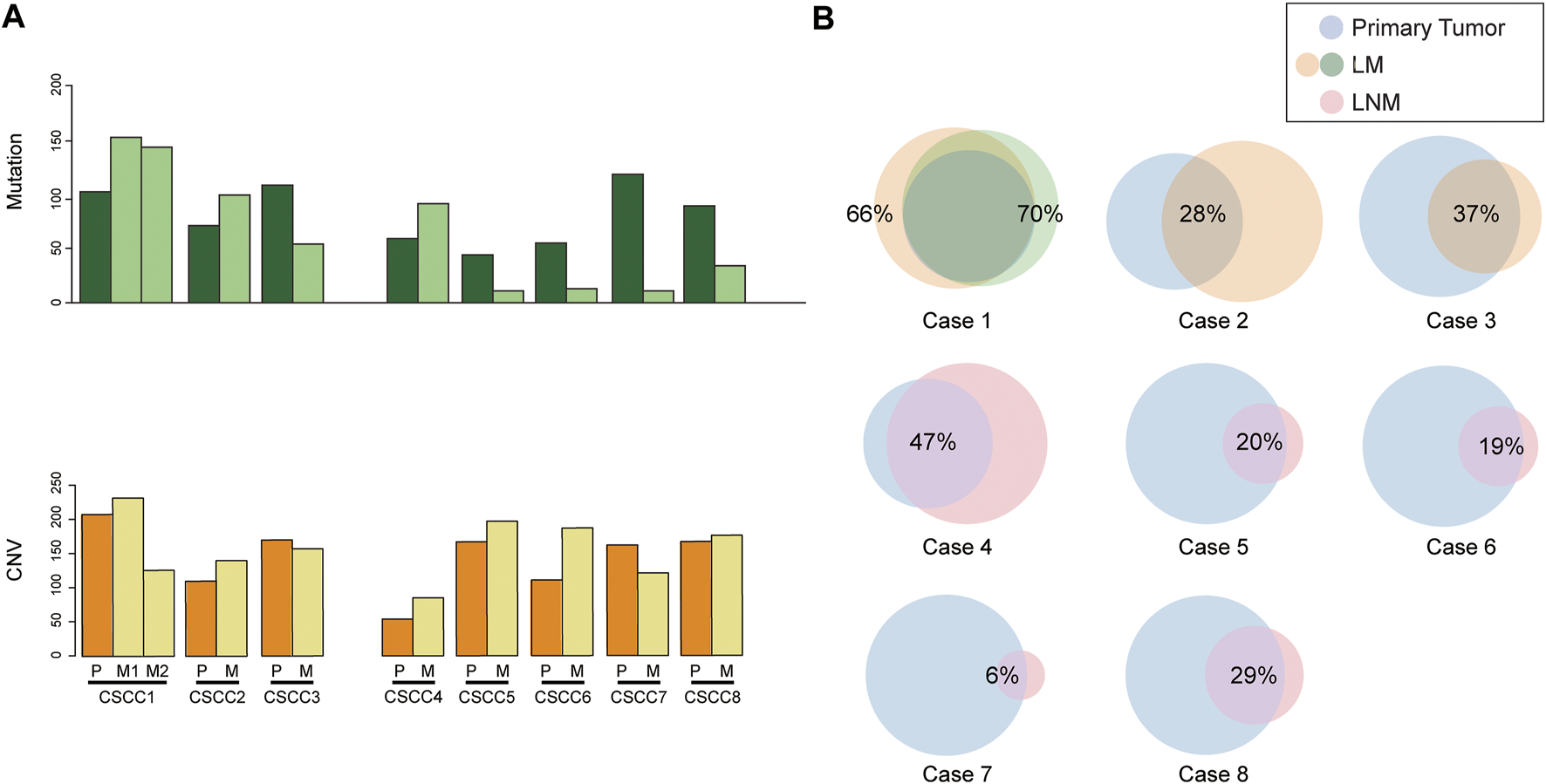
Comparison of somatic mutations in paired primary CSCC and metastatic CSCC. (A) mutation burden of eight CSCC pairs. (B) Venn diagrams depicting the overlap in somatic mutations in paired primary CSCC and metastatic CSCC. CSCC: cervical squamous cell carcinoma; P: primary lesions; M: metastatic lesions (lung metastasis in case 1–3; lymph nodes metastasis in case 4–8).
The chromosomal CNVs are a common phenomenon in CSCC [18, 19]. Genome-wide CNA profiling for eight pairs of primary CSCC and matched metastasis genomes are shown in Figure 2 and Supplementary Table S4. The number of CNVs is listed in Supplementary Table S5. CNVs were found to widely exist in primary and metastatic lesions. There was no statistical difference between the CNV number in LM and primary lesions (p = 0.593, Wilcoxon test). Similarly, there was no statistical difference in CNV between LNM lesions and primary lesions either (p = 0.225, Wilcoxon test). LM1 and LM2 in case 1 had distinct CNV numbers.
FIGURE 2
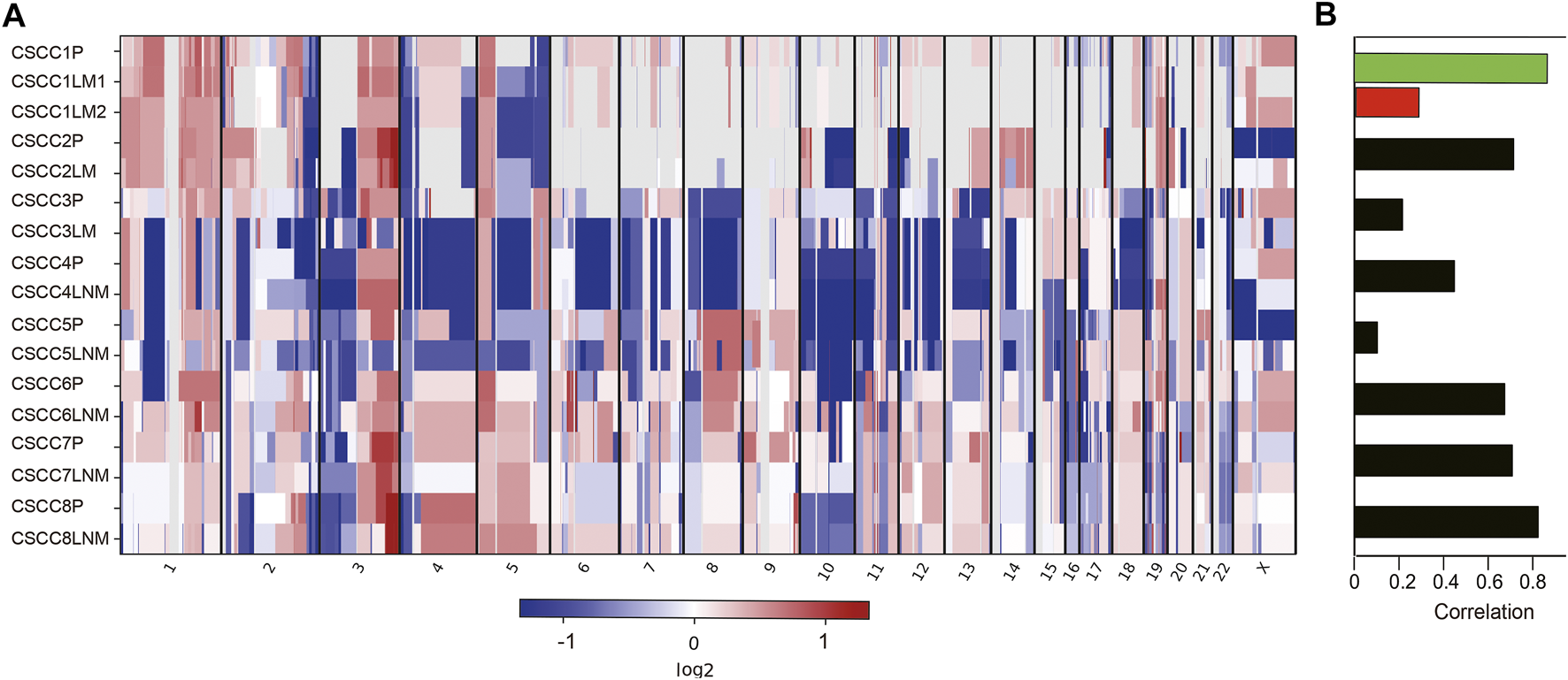
Somatic DNA copy number alterations in each CSCC patient. (A) Eight tumor samples were ordered from top to bottom in the chromosome plots, the Log2 ratios are plotted on the y-axis, and the genomic positions are plotted on the x-axis. (B) Spearman correlation between primary and matched metastatic CSCC. CSCC: cervical squamous cell carcinoma; P: primary lesions; LM: lung metastatic lesions; LNM: lymph nodes metastatic lesions.
Further analysis showed amplification of 9p CD274 and PDCD1LG2 (Programmed Cell Death 1 Ligand 2) were detected in the primary lesions of CSCC5 and CSCC7, and LNM lesion in CSCC6.
Whole-Exome Sequencing Shows Driver Gene Heterogeneity in Primary and Metastatic Specimens
Figure 3 and Supplementary Table S3 listed all cancer driver mutations identified in eight patients. The LNM in CSCC6 and CSCC7 did not harbor any driver mutations. The rest of the specimens harbored at least one cancer driver somatic mutation. The driver mutations in LNM were fewer than the primary lesions in all cases (p = 0.043, Wilcoxon test). The oncogenic tyrosine kinase pathway was frequently mutated; mutations involving KRAS and PTEN were detected in the primary specimen of CSCC4 and CSCC2. Gene mutations in the TGFβ signaling pathways could also be found, including CREBBP in the primary lesions of CSCC1 and CSCC7, the metastatic lesion in CSCC1.
FIGURE 3
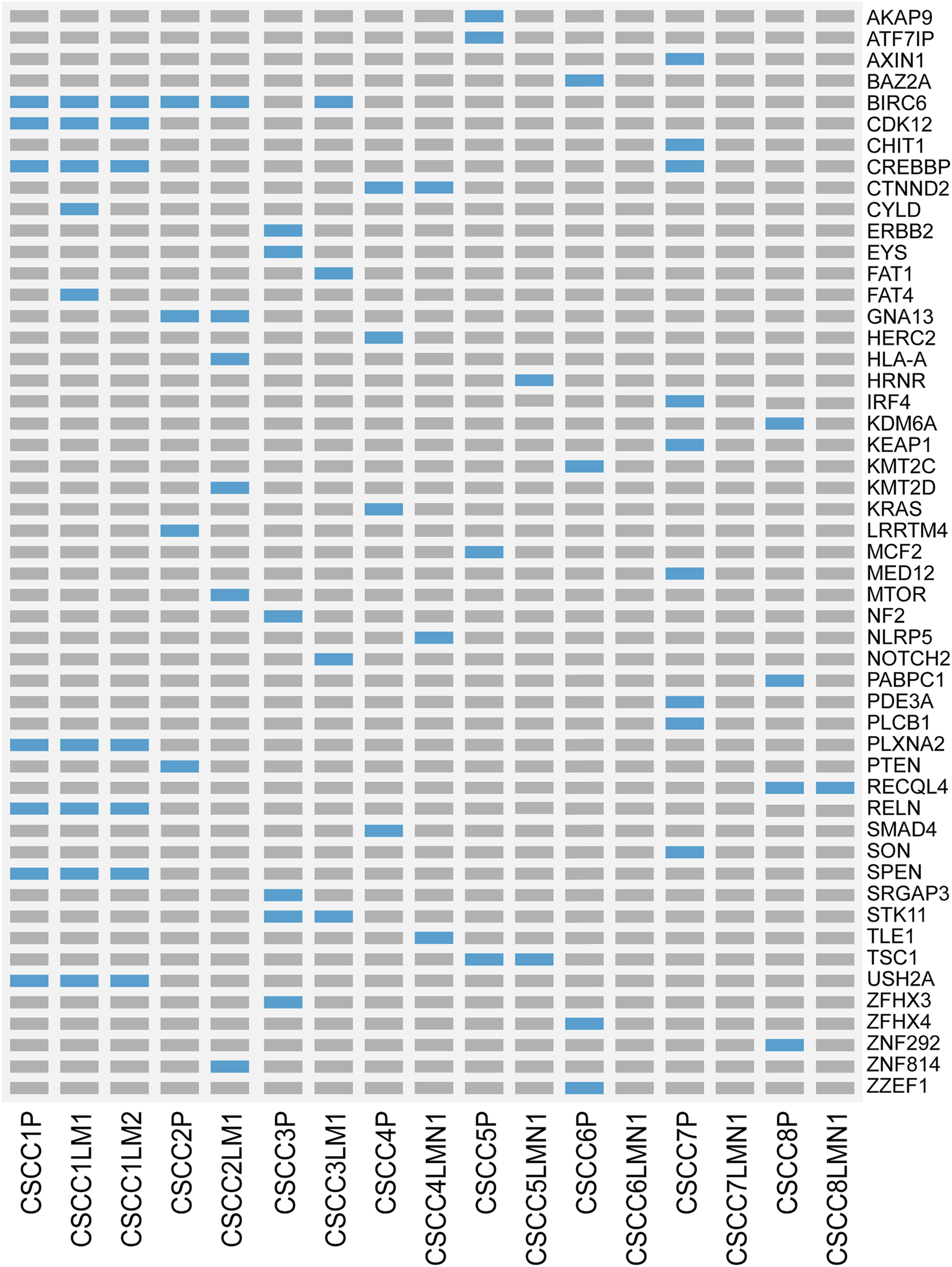
Landscape of somatic driver gene mutations in primary and metastatic CSCC. List of all cancer driver mutations in primary and metastatic CSCC. Light blue bars: single nucleotide variation; grey bars: no mutation was detected. CSCC: cervical squamous cell carcinoma; P: primary lesions; LM: lung metastatic lesions; LNM: lymph nodes metastatic lesions.
Whole-Exome Sequencing Shows the Clonal Relationship Between Primary and Metastatic Specimens
Figure 4 shows a phylogenetic tree of primary and metastatic lesions. The primary and metastatic lesions shared some mutations, similar to Figure 1B, which meant they were clonally related. And we could distinguish the LM from PPSCC. Further, we tried to develop a next-generation sequencing panel to help distinguish primary and metastatic pulmonary SCC. However, the genes were widely distributed. So, it was hard to apply a gene panel to distinguish the primary and metastatic pulmonary SCC.
FIGURE 4
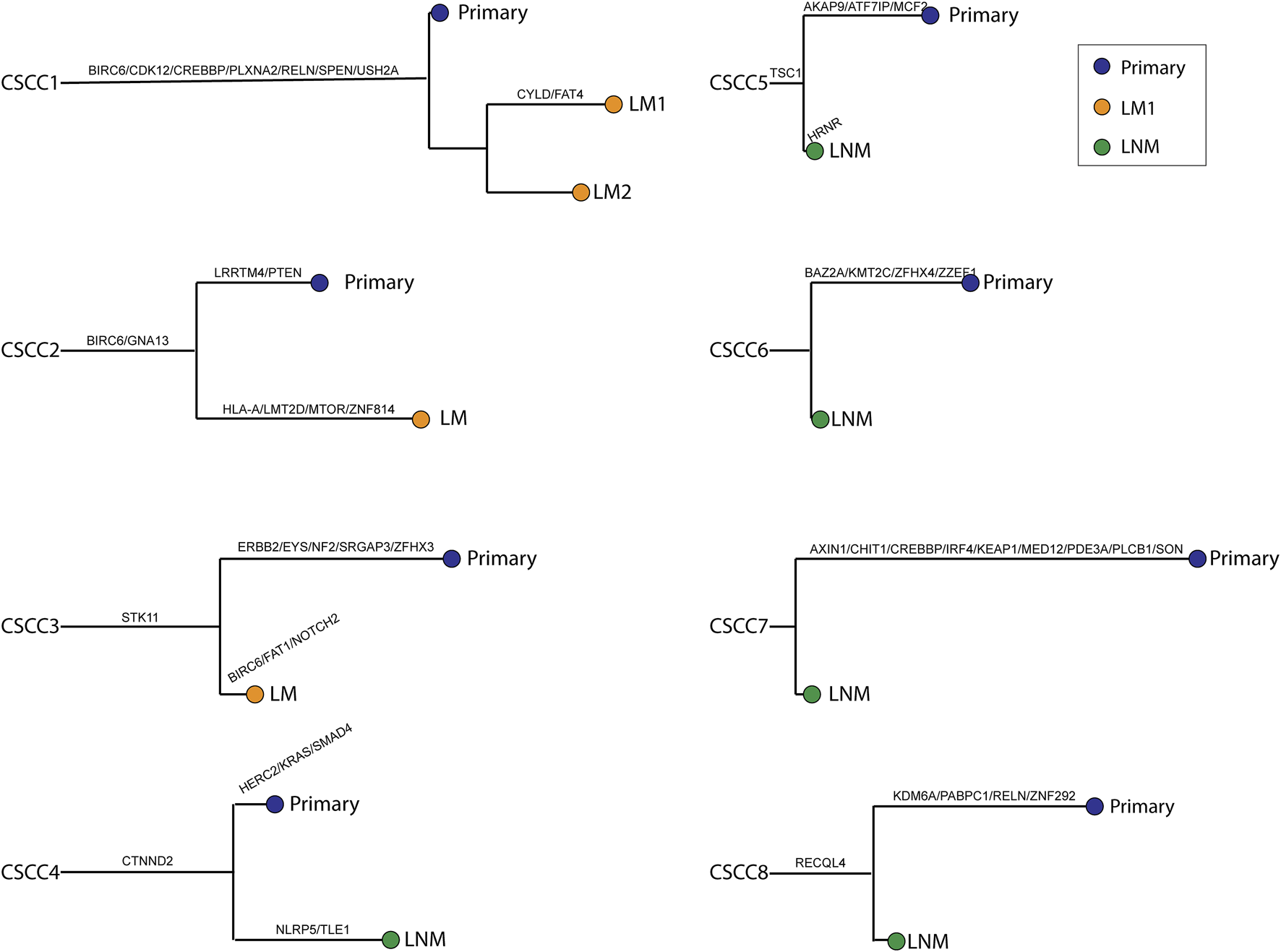
Phylogenetic trees were reconstructed for all cases. Branch length was relative to the number of non-synonymous somatic mutations (not only driver mutations); longer branches indicate more mutations. Branches are labeled with cancer-driver genes. The relative ordering of mutations is for visualization purposes only, as it is impossible to determine which came first. Each leaf corresponds to a single type of tissue harvested from the given patient. CSCC: cervical squamous cell carcinoma; P: primary lesions; LM: lung metastatic lesions; LNM: lymph nodes metastatic lesions.
Discussion
This study provides the first comprehensive analysis of the molecular genetic alterations in primary CSCC, LM, and LNM. Despite TCGA publishing the genetic landscape of cervical carcinoma, no genetic study on the metastatic CSCC has been published until now. With the deep sequencing of CSCC, we first established the tumor mutation burden and CNV difference on primary and metastatic CSCC.
The LM is originated from CSCC, which means they shared the original clone. The clones have some mutations, including driver mutations and passenger mutations, so the LM and CSCC shared some mutations. If we could find out the original mutations, or the mutations they shared, we could prove they are clonally related or have the same origin. First, we tried to develop a gene panel to include the common 100 genes of cervical carcinoma from NCBI and hope it could cover the >95% of cervical carcinoma cases, LM and LNM. Then we applied the panel to our data, but it could only distinguish six cases, so we expanded the panel to 500 genes, still failing to distinguish all the metastasis lesions. The first reason is that cervical carcinoma is a virus-driven cancer and the tumor mutations burden is low [20], which means their mutation quantity is low, therefore their shared mutations are pretty limited. The second reason is CSCC does not have specific mutations; the gene mutations are randomly distributed in different cases, therefore a panel could not cover all the cases. Thus, we concluded that WES but not an NGS (next-generation sequencing) gene panel is needed for distinguishing LM and PPSCC.
It is well-known that the accumulation of driver mutations [21] is needed for tumorigenesis, and at least three driver mutations are required for the tumorigenesis of pulmonary and colorectal cancers. Different from this, in this study, the LNM harbored fewer driver mutations than primary specimens and did not harbor any driver mutations in some cases. Thus, the metastasis clone expansion could be a distinct process compared to the tumor progression in primary lesions, it may not need as many driver mutations as the primary lesions. Meanwhile, because the driver mutations and somatic mutations in LNM were fewer than the primary CSCC, the tumor cells may migrate to lymph nodes in a very early stage. After the migration, the tumor cells in primary lesions and metastatic lesions have undergone separate clone development to harbor their private mutation as the phylogenetic tree showed (Figure 4). But the clone development in LNM may be slower than primary lesions because its mutations were fewer than the primary CSCC. Because of the early metastasis of LNM, we need to pay more attention to preventing LNM of early-stage CSCC.
Unlike LNM, LM harbored a similar number of driver mutations with primary lesions, so they may require sufficient driver mutations to expand clones. And the diagnosis for LM and primary CSCC has a time difference of 37 months on average, which means LM have a longer time to accumulate mutations than the primary lesions, but they harbored similar somatic and driver mutation number with primary CSCC so that the clone development may grow slower than the primary lesion because of the different microenvironment. But we do not rule out the possibility that the result is caused by the technical limitations and the involvement of DNA methylation.
From a genetics perspective, there must be mutations that convert primary cancer to a metastatic one, just as there are mutations that convert a normal cell to dysplasia or a carcinoma in situ to invasion. However, despite the intensive effort, consistent genetic alterations (including mutations and CNVs) that distinguish metastatic cancers from primary CSCC remain to be identified.
In a large study in 2017 [13], over 70% of cervical cancers exhibited genomic alterations in either one or both of PI3K–MAPK and TGFβ signaling pathways [13]. Like that result, mutations in PI3K–MAPK pathway and TGFβ signaling were detected in a significant percentage of patients in this study while the distribution of these mutations is not specific and could be found in both primary and metastatic lesions. But because of the pathway’s importance, it still needs in-depth research to clarify the role of PI3K–MAPK and TGFβ pathway in the process of oncogenesis in a more significant number of tumors.
One limitation of this study is the limited case number; a detailed sequence study should be performed on a larger cohort in the future. WES could detect SNVs and small indels, but could not recognize large indels, gene fusions, epigenetic changes, or post-translational, transcriptional dysregulations; this is another limitation of our study. Nonetheless, our results shed new light on the pathogenesis of tumor metastasis of CSCC, and our findings have implications for its early detection and prevention.
Key Messages
(1) Whole-exome sequencing is a helpful tool in distinguishing primary pulmonary squamous cell carcinoma and metastatic cervical squamous cell carcinoma (CSCC).
(2) The lymph nodes metastasis harbored fewer cancer driver mutations than primary CSCC specimens with a significant difference.
(3) There was no significant difference in somatic mutations and copy number variation (CNV) between primary and metastatic CSCC.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving human participants were reviewed and approved by the Institutional Review Board of Cancer Hospital, Chinese Academy of Medical Sciences. The patients/participants provided their written informed consent to participate in this study.
Author contributions
LL and QS contributed equally. YJ, GY and YS conceived the study, and LL, QS and DC carried out experiments. LL, QS, and DC were involved in analyzing the data. LL, QS and YS were involved in interpreting the data. LL and YS wrote the paper. GY and YS are co-corresponding authors.
Funding
This work was supported by the CAMS Innovation Fund for Medical Sciences (CIFMS) (grant numbers 2017-I2M-3-020); the Beijing Hope Run Special Fund of Cancer Foundation of China (grant numbers LC2017A10, LC2020A08, and LC2020B21).
Conflict of interest
QS and DC were employed by the Genetron Health (Beijing) Co. Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.por-journal.com/articles/10.3389/pore.2022.1610325/full#supplementary-material
Supplementary Figure S1Photomicrographs from a representative case (case 2).
References
1.
Siegel RL Miller KD Fuchs HE Jemal A . Cancer Statistics, 2021. CA A Cancer J Clin (2021) 71(1):7–33. 10.3322/caac.21654
2.
Walboomers JMM Jacobs MV Manos MM Bosch FX Kummer JA Shah KV et al Human Papillomavirus Is a Necessary Cause of Invasive Cervical Cancer Worldwide. J Pathol (1999) 189(1):12–9. 10.1002/(sici)1096-9896(199909)189:1<12::aid-path431>3.0.co;2-f
3.
Fulcher AS O'Sullivan SG Segreti EM Kavanagh BD . Recurrent Cervical Carcinoma: Typical and Atypical Manifestations. Radiographics (1999) 19:S103–S116. 10.1148/radiographics.19.suppl_1.g99oc19s103
4.
Ki EY Lee KH Park JS Hur SY A Clinicopathological Review of Pulmonary Metastasis from Uterine Cervical Cancer. Cancer Res Treat (2016) 48(1):266-72
5.
Chang SY Keeney M Law M Donovan J Aubry M-C Garcia J . Detection of Human Papillomavirus in Non-Small Cell Carcinoma of the Lung. Hum Pathol (2015) 46(11):1592–7. 10.1016/j.humpath.2015.07.012
6.
Lin S Zhang X Li X Qin C Zhang L Lu J et al Detection of Human Papillomavirus Distinguishes Second Primary Tumors from Lung Metastases in Patients with Squamous Cell Carcinoma of the Cervix. Thorac Cancer (2020) 11(8):2297–305. 10.1111/1759-7714.13544
7.
Fan X Yu K Wu J Shao J Zhu L Zhang J . Correlation Between Squamous Cell Carcinoma of the Lung and Human Papillomavirus Infection and the Relationship to Expression of P53 and P16. Tumor Biol (2015) 36(4):3043–9. 10.1007/s13277-014-2940-7
8.
Weichert W Schewe C Denkert C Morawietz L Dietel M Petersen I . Molecular HPV Typing as a Diagnostic Tool to Discriminate Primary from Metastatic Squamous Cell Carcinoma of the Lung. Am J Surg Pathol (2009) 33(4):513–20. 10.1097/PAS.0b013e3181938319
9.
Crook T Wrede D Tidy JA Vousden KH Wrede D Tidy J et al Clonal P53 Mutation in Primary Cervical Cancer: Association with Human-Papillomavirus-Negative Tumours. The Lancet (1992) 339(8801):1070–3. 10.1016/0140-6736(92)90662-m
10.
McIntyre JB Wu JS Craighead PS Phan T Köbel M Lees-Miller SP et al PIK3CA Mutational Status and Overall Survival in Patients with Cervical Cancer Treated with Radical Chemoradiotherapy. Gynecol Oncol (2013) 128(3):409–14. 10.1016/j.ygyno.2012.12.019
11.
Kang S Kim H-S Seo SS Park S-Y Sidransky D Dong SM . Inverse Correlation Between RASSF1A Hypermethylation, KRAS and BRAF Mutations in Cervical Adenocarcinoma. Gynecol Oncol (2007) 105(3):662–6. 10.1016/j.ygyno.2007.01.045
12.
Wingo SN Gallardo TD Akbay EA Liang M-C Contreras CM Boren T et al Somatic LKB1 Mutations Promote Cervical Cancer Progression. PLoS One (2009) 4(4);e5137. 10.1371/journal.pone.0005137
13.
Cancer Genome Atlas Research N Albert Einstein College of M . Integrated Genomic and Molecular Characterization of Cervical Cancer. Nature (2017) 543(7645):378–84. 10.1038/nature21386
14.
Dietlein F Weghorn D Taylor-Weiner A Richters A Reardon B Liu D et al Identification of Cancer Driver Genes Based on Nucleotide Context. Nat Genet (2020) 52(2):208–18. 10.1038/s41588-019-0572-y
15.
Robinson JT Thorvaldsdóttir H Winckler W Guttman M Lander ES Getz G et al Integrative Genomics Viewer. Nat Biotechnol (2011) 29(1):24–6. 10.1038/nbt.1754
16.
Talevich E Shain AH Botton T Bastian BC . CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. Plos Comput Biol (2016) 12(4);e1004873. 10.1371/journal.pcbi.1004873
17.
Phylip . Phylogeny Inference Package) Version 3.6. WA: Department of Genome Sciences, University of Washington, Seattle. (2015).
18.
Bodelon C Vinokurova S Sampson JN den Boon JA Walker JL Horswill MA et al Chromosomal Copy Number Alterations and HPV Integration in Cervical Precancer and Invasive Cancer. Carcin (2016) 37(2):188–96. 10.1093/carcin/bgv171
19.
Yan D Yi S Chiu WC Qin LG Kin WH Kwok Hung CT et al Integrated Analysis of Chromosome Copy Number Variation and Gene Expression in Cervical Carcinoma. Oncotarget (2017) 8(65):108912–22. 10.18632/oncotarget.22403
20.
Wang M Fan W Ye M Tian C Zhao L Wang J et al Molecular Profiles and Tumor Mutational Burden Analysis in Chinese Patients with Gynecologic Cancers. Sci Rep (2018) 8(1):8990. 10.1038/s41598-018-25583-6
21.
Tomasetti CC Marchionni MA Nowak B Parmigiani G Vogelstein B . Only Three Driver Gene Mutations Are Required for the Development of Lung and Colorectal Cancers. Proc Natl Acad Sci U.S.A (2015) 112(1):118–23. 10.1073/pnas.1421839112
Summary
Keywords
metastasis, cervix, squamous cell carcinoma, sequence, lung
Citation
Li L, Song Q, Cao D, Jiao Y, Yuan G and Song Y (2022) Whole-Exome Sequencing Could Distinguish Primary Pulmonary Squamous Cell Carcinoma From Lung Metastases in Individuals With Cervical Squamous Cell Carcinoma. Pathol. Oncol. Res. 28:1610325. doi: 10.3389/pore.2022.1610325
Received
20 January 2022
Accepted
30 March 2022
Published
11 May 2022
Volume
28 - 2022
Edited by
Natalia Buza, Yale University, United States
Updates
Copyright
© 2022 Li, Song, Cao, Jiao, Yuan and Song.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Song, songyan@cicams.ac.cn; Guangwen Yuan, william327@126.com
†These authors have contributed equally to this work
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.