- 1Department of Surgical Pathology, Hyogo College of Medicine, Nishinomiya, Japan
- 2Departtment of Gastroenterological Surgery, Osaka University Graduate School of Medicine, Suita, Japan
- 3Japan Community Healthcare Organization (JCHO) Osaka Hospital, Osaka, Japan
- 4Department of Pathology, Faculty of Medicine, Kindai University, Osaka, Japan
Gastrointestinal stromal tumor (GIST), the most common mesenchymal tumor of the human gastrointestinal tract, differentiating toward the interstitial cell of Cajal (ICC), arises predominantly in the stomach and small intestine. Small intestinal GISTs appear to have worse prognosis than gastric GISTs. In a pilot study of a cDNA expression chip using several GISTs, we found that Cell Adhesion Molecule 1 (CADM1), which could contribute to tumor growth and infiltration, is expressed more strongly in small intestinal GISTs than gastric GISTs. In the present study, we examined CADM1 expression in GISTs of different sites and with different gene abnormalities using a large number of gastric and small intestinal GISTs. First, immunoblotting confirmed significantly higher CADM1 expression in small intestinal GISTs with exon 11 c-kit mutation than gastric GISTs with exon 11 c-kit mutation. Real-time PCR also revealed that small intestinal GISTs with exon 11 c-kit mutation showed significantly higher CADM1 mRNA than gastric GISTs with exon 11 c-kit mutation. Although most small intestinal GISTs showed high CADM1 mRNA expression regardless of gene abnormality types, different CADM1 expression was detected between gastric GISTs with c-kit mutation and those with PDGFRA mutation. Immunohistochemistry showed that many small intestinal GISTs were CADM1-positive but most gastric GISTs CADM1-negative or -indefinite. In the normal gastric and small intestinal walls, immunoreactivity of CADM1 was detected only in nerves, but neither in gastric ICCs nor small intestinal ICCs, indicating that the high CADM1expression in small intestinal GISTs might be acquired during tumorigenesis. Different CADM1 expression between gastric and small intestinal GISTs might be related to different prognoses between them. Further functional experiments are needed to elucidate the role of CADM1 on GIST biology, and there is a possibility that targeting therapy against CADM1 has a preventive effect for tumor spreading in small intestinal GISTs.
Introduction
Gastrointestinal stromal tumor (GIST) is the most common mesenchymal tumor of the human gastrointestinal tract, differentiating toward the interstitial cell of Cajal (ICC) [1]. Sporadic GISTs usually harbor gain-of-function mutations in either c-kit (80–85%) or PDGFRA (5–10%) [2]. Most of the c-kit mutations in GISTs are present at exon 11, but a minority at exon 8, 9, 13 or 17 [3]. Characteristically, most GISTs with exon 9 c-kit mutations are located in the small intestine [4]. On the other hand, nearly all GISTs with PDGFRA mutations occur in the stomach. Approximately 10% of GISTs, defined as c-kit/PDGFRA wild-type (WT) GISTs [5], which carry wild-type sequences in all hot spots of c-kit and PDGFRA, represent some groups with distinct molecular characteristics, including defects in SDH complex [6], mutations of NF1 [7], or mutations of BRAF [8].
GISTs show a wide spectrum of biologic behavior ranging from benign to malignant, varying by tumor size, mitotic activity and anatomical site. GISTs arise predominantly in the stomach (60–70%) and small intestine (20–30%) [9]. Some studies have suggested that the anatomical site has a prognostic value independent of tumor size and mitotic counts [10, 11]. Small intestinal GISTs usually show a higher malignant potential than gastric GISTs [11–13]. Distinct transcription profiles related to the anatomical location in GISTs have been reported [14, 15]. Hierarchical clustering analysis of the transcripts shows that GISTs are roughly separated into two groups such as gastric GISTs and small intestinal GISTs, and the result may partially explain why GISTs of the small intestine show more aggressive behavior than those of the stomach, with similar size and mitotic rate.
Cell adhesion molecule 1 (CADM1) is a member of the immunoglobulin superfamily initially known as spermatogenic immunoglobulin superfamily (SgIGSF) and synaptic cell adhesion molecule (SynCAM) [16–18]. CADM1 mediates formation of synapses between neurons, and adhesion between mast cells and nerve [17, 19], fibroblast [20], extracellular matrix [21] and muscular cells [19, 20] either homophilically or heterophilically. Moreover, CADM1 plays a role in adhesion between spermatogonia and Sertoli cells [22], or islet cells [23]. CADM1 was also identified as a tumor suppressor of lung cancer 1 (TSLC1) [24]. Loss of CADM1 expression due to methylation was frequently found in many kinds of epithelial cancer [25], such as gastric cancer [26], breast cancer [27, 28], and squamous cell carcinoma [29–31]. In contrast, CADM1 is upregulated in adult T cell leukemia and acute myelocytic leukemia, contributing to the enhancement of cell-cell adhesion to the vascular endothelium, tumor growth and tissue infiltration [32, 33].
In a pilot study using a small number of GIST cases, we found that small intestinal GISTs showed stronger expression of CADM1 mRNA than gastric GISTs (data not shown). Here we assessed the expression of CADM1 at the protein and mRNA levels using a large number of gastric and small intestinal GISTs to clarify whether higher CADM1 expression in small intestinal GISTs than gastric GISTs is really common and whether the expression of CADM1 is different in GISTs with different types of gene abnormalities. We also performed immunofluorescent staining of CADM1 on normal gastric and small intestinal ICCs to examine whether the distinct expression of CADM1 in GISTs at different anatomical sites is related to that in ICCs at different anatomical sites.
Materials and Methods
Tissue Specimens
In total, 107 cases previously diagnosed as GIST by pathological examination including KIT immunohistochemistry and c-kit/PDGFRA gene analyses were chosen for the present study. They included 76 fresh frozen GIST tissues (Table 1) and the other 31 formalin-fixed paraffin-embedded (FFPE) GIST tissues. Seventy-six fresh frozen GIST tissues from August 1995 to August 2016 at Hyogo College of Medicine Hospital and Osaka University Medical Hospital were used for Western blotting (16 cases) and real-time PCR experiments (62 cases) (Two cases were overlapped). Of 76 fresh frozen GISTs, 36 (47.4%) were duodenal/jejuno-ileal ones and 40 (52.6%) were gastric ones (Table 1). Of 36 duodenal/jejuno-ileal GISTs, 21 (58.3%) had exon 11 c-kit mutation, 10 (27.8%) had exon 9 c-kit mutation, and 5 (13.9%) had NF1 abnormality (Table 1). Of 40 gastric GISTs, on the other hand, 21 (52.5%) had exon 11 c-kit mutation, 5 (12.5%) had exon 17 c-kit mutation and 14 (35.0%) had exon 18 PDGFRA mutation (Table 1). Tumor size, mitotic rate and clinical outcome of them were also shown in Table 1. Thirty-one cases of FFPE GIST tissues including 15 jejuno-ileal ones and 16 gastric ones were from Hyogo College of Medicine Hospital between 2007 and 2018 were used for immunohistochemistry. For CADM1 cDNA sequencing, 10 duodenal/jejuno-ileal GISTs among ones served for real-time PCR were used.
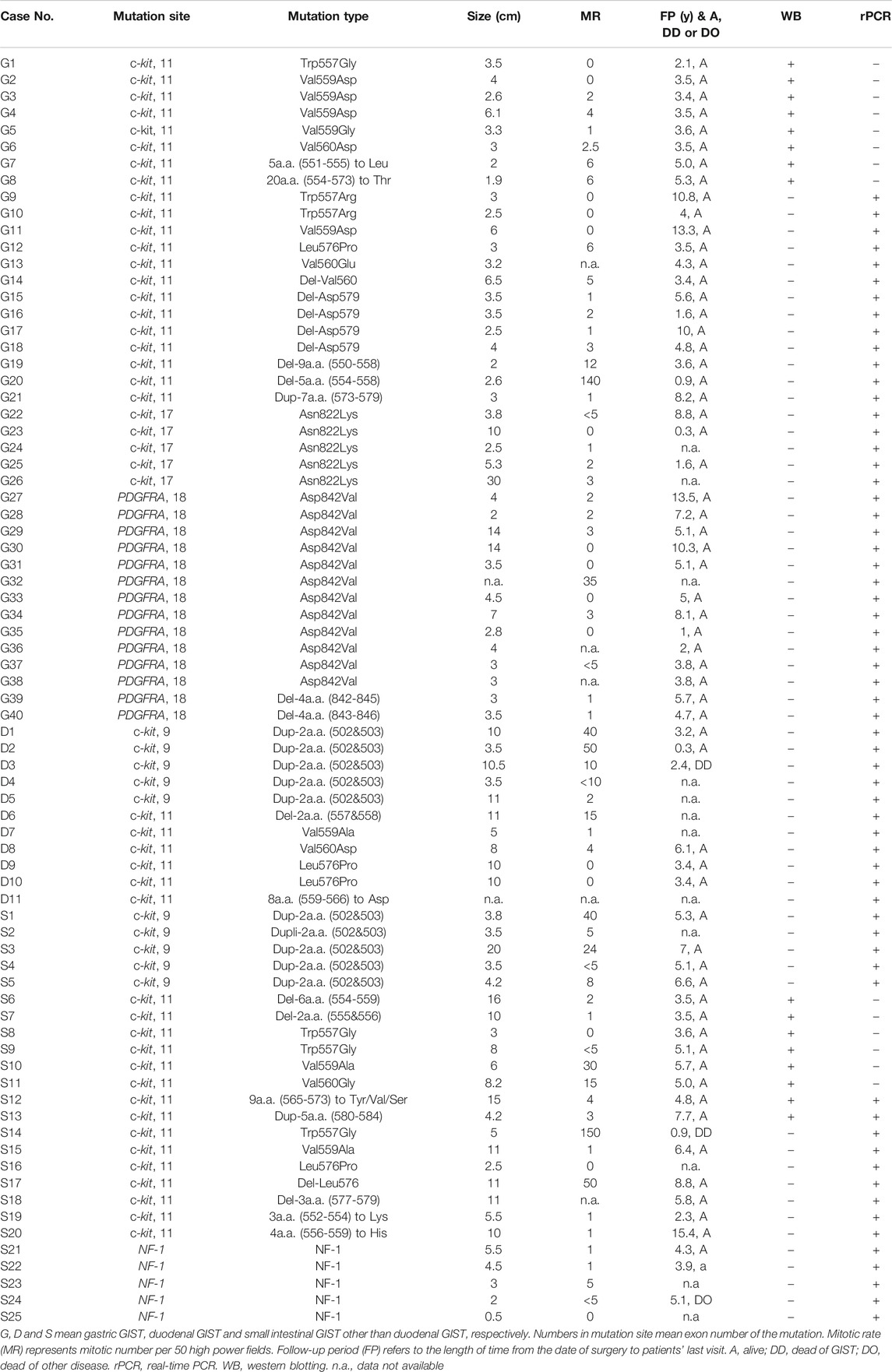
TABLE 1. Clinicopathological Characteristics of gastric and duodenal/jejuno-ileal GISTs examined for real-time PCR and Western blotting.
Western Blotting
Equals of protein samples extracted from fresh frozen tissues of eight small intestinal (jejuno-ileal) GISTs with exon 11 c-kit mutation and eight gastric GISTs with exon 11 c-kit mutation were separated by 12% SDS-PAGE and subsequently transferred to PVDF membrane (Invitrogen, Carlsbad, CA). Membrane was incubated with anti-CADM1 chicken monoclonal antibody (MBL International, Woburn, MA) diluted at 1:1,000 for 3 h at room temperature using iBindTM Western System (Invitrogen). Immunoreactive protein bands were visualized using Prime Western Blotting Detection Reagent (GE Healthcare Life Science, Buckinghamshire, United Kingdom) on ChemiDoc Imaging System (Bio-Rad, Laboratories, California, United States). Reprobing by anti-KIT rabbit polyclonal antibody (DAKO, Carpinteria, CA) and anti-β-actin mouse monoclonal antibody (Abcam, Cambridge, United Kingdom) was done.
Real-Time Quantitative RT-PCR
Total RNA was extracted from 62 fresh frozen GIST tissues using RNeasy Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s recommendation. Complementary DNA was synthesized from total RNA (10 μg) using reverse transcriptase (Invitrogen). Real-time PCR analysis was performed using the Applied Biosystems STEP ONETM standard real-time PCR system (Applied Biosystems, Foster City, CA) based on published sequences for genes encoding the human CADM1, c-kit, and GAPDH (Applied Biosystems). For each sample, total volume of 20 μl consisted of 10 μl TaqMan Gene Expression Master Mix (2x, Applied Biosystems), 1 μl TaqMan Probe, 1 μl of each cDNA template and 8 μl of distilled water. The real-time PCR fragments were amplified as follows: one cycle at 48°C for 30 min, 40 cycles at 95°C for 10 min and 60°C for 1 min. Expression level of CADM1 mRNA and c-kit mRNA was standardized by expression of GAPDH mRNA.
Immunohistochemistry
Three μm-thick sections were cut from formalin-fixed and paraffin embedded tissues, mounted on slides coated with 3-aminopropyltriethoxysilane, deparaffinized in xylene and rehydrated in descending grades of ethanol. Antigen retrieval was performed in a water bath for 20 min at 98°C at Target Retrieval Solution, pH 9.0 (DAKO). Nonspecific staining was blocked by treating the sections with 5% donkey serum in Tris-buffered saline (pH 6.0) for 30 min. CADM1 immunohistochemistry was performed with a monoclonal chicken antibody (3E1, MBL International) diluted at 1:1,000 in Antibody Diluent (DAKO). The slides were incubated with donkey anti-chicken IgY-HRP conjugated (EMD Millipore, Temecula, CA) as a secondary antibody at 1:500 dilution. Signals were developed with diaminobenzidine substrate chromogen system (DAKO), and counterstained with hematoxylin. Images were obtained under All-in-One Fluorescence Microscope (KEYENCE, Osaka, Japan).
Immunofluorescence Staining
Deparaffinization and rehydration of the sections were performed as described. After the sections were autoclaved for 20 min at 121°C in Target Retrieval Solution, pH 9.0 (DAKO), they were doubly immunostained with a combination of the anti-CADM1 antibody (MBL International) diluted at 1:1,000 and anti-c-kit antibody (NovocastraTM) diluted at 1:100 for 1 h in room temperature, followed by Cy5-conjugated anti-chicken IgY and Cy2-conjugated anti-rabbit IgG secondary antibodies (Jackson ImmunoResearch, West Grove, PA), respectively in dark for 45 min. The slides then mounted with antifade mountant with DAPI (GE Healthcare Life Science), and examined with a laser confocal scanning system (ZEISS, LSM780, Oberkochen, Germany) under 510 and 670 nm.
cDNA Sequencing of CADM1
Total RNA extracted from 10 fresh small intestinal GIST tissues with high CADM1 expression was reverse-transcribed to cDNA, and PCR for CADM1 cDNA amplification was performed. Primer sequences are listed in Supplementary Table S1. PCR products were purified with the Qiaquick Gel Extraction Kit (QIAGEN) and sequenced on both strands using Big Dye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems) and ABI 3100-Avant Genetic Analyzer (Applied Biosystems).
Statistical Analysis
Statistical analyses were performed using Prism GraphPad 8 (GraphPad Software, La Jolla, CA). Two-tailed unpaired Student’s t-tests with Welch’s correction or one-way ANOVA for multiple comparisons when appropriate were used to determine statistical significance (p < 0.05 was considered statistically significant).
Results
Expression of CADM1 Protein in Gastric and Small Intestinal GISTs with Exon 11 c-Kit Mutation Examined by Western Blotting
CADM1 expression at the protein level was assessed in eight small intestinal (jejuno-ileal) and eight gastric GISTs with exon 11 c-kit mutation (Table 1) by Western blotting. Apparent bands of CADM1 protein were detected in all small intestinal GISTs with exon 11 c-kit mutation but not in all gastric GISTs with exon 11 c-kit mutation (Figure 1). All small intestinal and gastric GISTs with exon 11 c-kit mutation examined strongly expressed KIT protein (Figure 1).
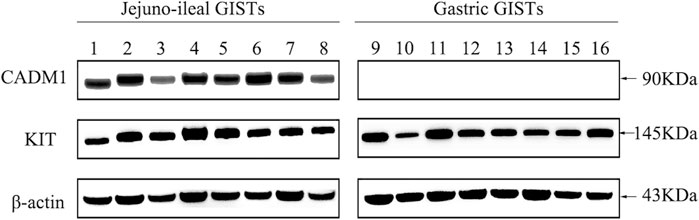
FIGURE 1. Distinct expression of CADM1 protein between small intestinal and gastric GISTs with exon 11 c-kit mutation by Western blotting. CADM1 protein was abundantly detected in all cases (n = 8) of small intestinal (jejuno-ileal) GISTs with exon 11 c-kit mutation, while that was not apparent in all gastric GISTs with exon 11 c-kit mutation (n = 8). All jejuno-ileal and gastric GISTs with exon 11 c-kit mutation examined strongly expressed KIT protein, and expression of β-actin as a control was normally detected in all samples.
Expression of CADM1 mRNA in Gastric and Small Intestinal GISTs with Various Gene Abnormalities Examined by Real-Time PCR
CADM1 mRNA expression was quantified by real-time PCR in gastric and small intestinal GISTs with various gene abnormalities. First, we compared expression level of CADM1 mRNA in duodenal/jejuno-ileal GISTs with exon 11 c-kit mutation and that in gastric GISTs with exon 11 c-kit mutation (Table1). Significantly higher expression of CADM1 mRNA was observed in 15 duodenal/jejuno-ileal GISTs with exon 11 c-kit mutation than in 13 gastric GISTs with exon 11 c-kit mutation (p < 0.0001, Figure 2), consistent with the protein analysis (Figure 1). Next, expression level of CADM1 mRNA was compared among duodenal/jejuno-ileal GISTs with various gene abnormalities (Table 1). Ten duodenal/jejuno-ileal GISTs with exon 9 c-kit mutation and five those from NF1 patients (Table 1) showed high expression of CADM1 mRNA, similar to those with exon 11 c-kit mutation (Figure 3A). All types of duodenal/jejuno-ileal GISTs with different gene abnormalities also showed similar levels of c-kit mRNA (Figure 3B). Then, we compared expression level of CADM1 mRNA among gastric GISTs with various gene abnormalities (Table 1). Fourteen gastric GISTs with exon 18 PDGFRA mutation expressed significantly higher mRNA of CADM1 than 13 those with exon 11 c-kit mutation (p = 0.0028) and five those with exon 17 c-kit mutation (p = 0.0403) (Table 1; Figure 3C), but the level of CADM1 mRNA expression even in gastric GISTs with exon 18 PDGFRA mutation was significantly lower than that in small intestinal GISTs with various types of gene abnormalities (p = 0.0005) (Figures 3A,C). Expression of c-kit mRNA in gastric GISTs with exon 11 and exon 17 c-kit mutations was significantly higher (p = 0.0004 and p = 0.0200, respectively) than that in small intestinal GISTs with exon 11 c-kit mutation (Figures 3B,D), but that in gastric GISTs with exon 18 PDGFRA mutation was significantly lower than that in gastric GISTs with exon 11 and exon 17 c-kit mutation (p < 0.0001 and p < 0.0001, respectively) (Figure 3D).
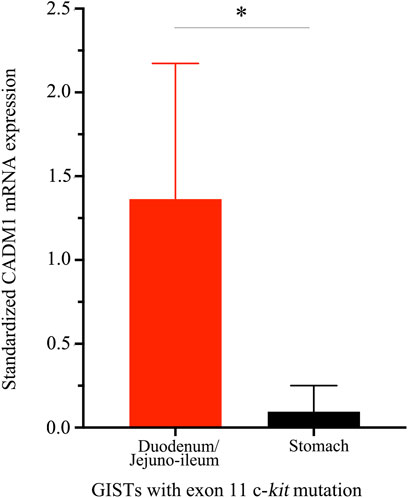
FIGURE 2. Distinct CADM1 mRNA levels quantified by real-time PCR between small intestinal and gastric GISTs with exon 11 c-kit mutation. Quantitative RT-PCR demonstrated that level of CADM1 mRNA expression standardized by GAPDH mRNA expression in small intestinal GISTs with exon 11 c-kit mutation (n = 15) and that in the gastric GISTs with exon 11 c-kit mutation (n = 13) was statistically significant (*p < 0.0001).
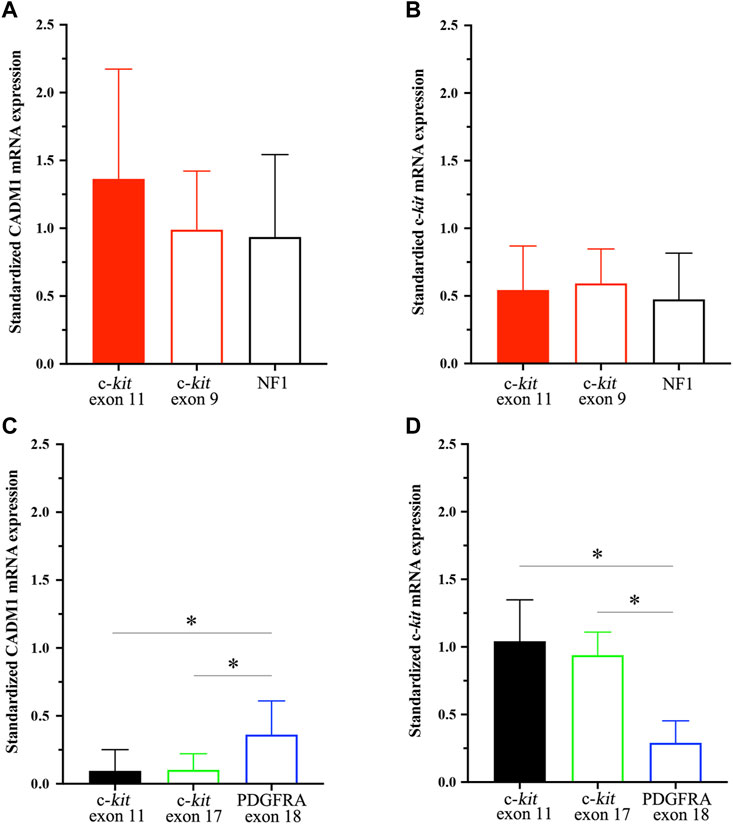
FIGURE 3. CADM1 and c-kit mRNA levels quantified by real-time PCR in small intestinal GISTs and gastric GISTs with different gene abnormalities. (A). CADM1 mRNA expression level standardized by GAPDH mRNA expression showed no significant difference among small intestinal GISTs with exon 11 c-kit mutation, those with exon 9 c-kit mutation and those from NF1 patients. (B). Expression level of c-kit mRNA standardized by GAPDH mRNA expression was similar among small intestinal GISTs with exon 11 c-kit mutation, those with exon 9 c-kit mutation and those from NF1 patients. (C). CADM1 mRNA expression level standardized by GAPDH mRNA expression in gastric GISTs with exon 18 PDGFRA mutation was significantly higher than that with exon 11 c-kit mutation (p = 0.0028) or exon 17 c-kit mutation (p = 0.0403). (D). Expression level of c-kit mRNA standardized by GAPDH mRNA expression in gastric GISTs with exon 18 PDGFRA mutation was significantly lower than that with exon 11 or exon 17 c-kit mutation (p < 0.001). * means statistical significance (p < 0.05).
Immunohistochemical and Immunofluorescent Staining of CADM1 in Gastric and Small Intestinal GISTs
To confirm whether CADM1 protein is expressed by tumor cells themselves of small intestinal GISTs, immunohistochemistry for CADM1 was performed. According to membranous staining pattern, CADM1 expression was divided into three categories; no staining (negative) (Figure 4A), weak staining (indefinite) (Figure 4B) and strong staining (positive) (Figure 4C). Diffuse and strong immunoreactivity (positive) for anti-CADM1 was observed in 11 cases (73.3%) of small intestinal GISTs, but the others had no staining (negative) in 2 cases or weak staining (indefinite) in 2 cases. Only 1 of 16 (6.25%) gastric GISTs showed focal positive staining for CADM1, 9 no staining (negative), and six weak staining (indefinite). Immunofluorescent staining of CADM1 (Figure 5A) and KIT (Figure 5B) in small intestinal GISTs clearly showed double positivity of them (Figure 5C). On the other hand, gastric GISTs showed KIT expression (Figures 5E,F) but not CADM1 expression (Figures 5D,F).
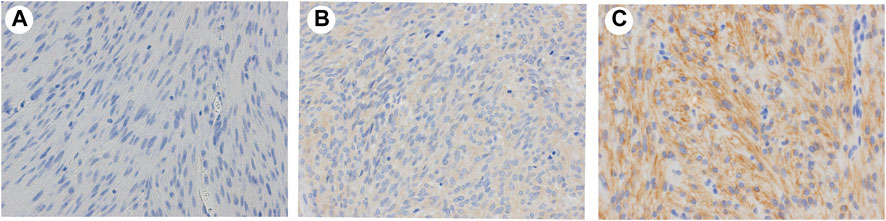
FIGURE 4. Representative results of expression of CADM1 immunostaining in GISTs. (A). Negative: no apparent CADM1 staining. (B). Indefinite: weak CADM1 cytoplasmic staining but no membranous staining. (C). Positive: diffuse or focal strong CADM1 membranous staining. (A). Gastric GIST. (B) and (C). Small intestinal GISTs. Original magnification: ×400.
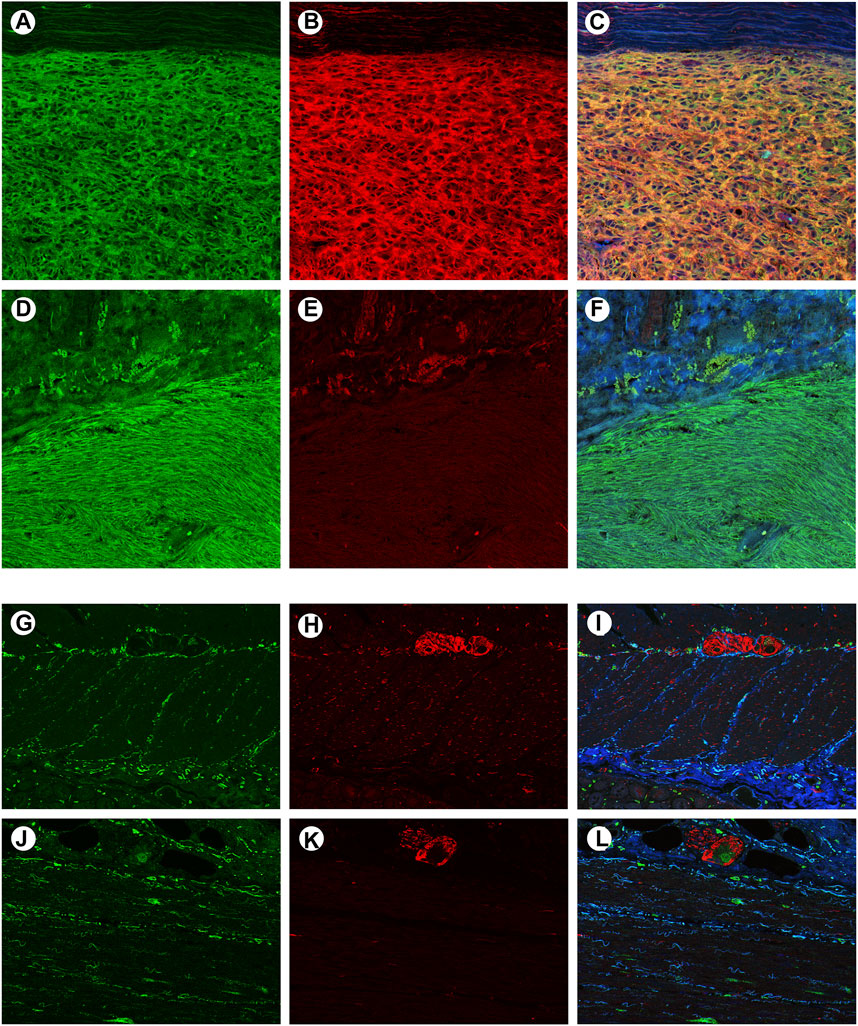
FIGURE 5. Confocal microscopic images of immunofluorescent staining with KIT and CADM1 in a representative small intestinal GIST, a representative gastric GIST, normal small intestinal wall and normal gastric wall. (A). The small intestinal GIST was stained by KIT (green). (B). The small intestinal GIST was also stained by CADM1 (red), (C). Merged image of panels “a” and “b” showed yellow color, indicating co-expression of c-kit and CADM1 by the tumor tissue. (D). The gastric GIST was stained with KIT (green). (E). No apparent staining with CADM1 was detected in the gastric GIST. (F). Merged image of panels “d” and “e” was showed only green color. (G). KIT-positive cells (green) were located around the myenteric plexus and within the muscular layers in small intestinal wall. (H). Immunoreactivity for CADM1 (red) was only observed in nerve strands. (I). Merged image of KIT and CADM1 immunoreactivities showed no doubly stained cells in small intestinal wall. (J). KIT-positive cells (green) were also detected around the myenteric plexus in gastric wall. (K). CADM1-positive cells (red) were only seen in nerve tissues. l. The merged image of KIT and CADM1 immunoreactivities did not show double positive cells in gastric wall. a-l. Original magnification: ×200.
Immunofluorescence Staining of CADM1 in Normal Gastric and Small Intestinal Walls
To clarify the localization of CADM1 protein in normal gastrointestinal tract, double immunofluorescence staining of KIT and CADM1 was performed in normal small intestinal and gastric walls. In the small intestinal wall, KIT-positive ICCs were located around the myenteric plexus and in the muscular layers (Figure 5G). On the other hand, immunoreactivity for CADM1 was only observed at nerve strands (Figure 5H). Thus, the merged image of KIT immunoreactivity and CADM1 immunoreactivity showed no doubly stained cells (Figure 5I). In the wall of the stomach, KIT-positive ICCs were also observed around the myenteric plexus and in the muscular layers (Figure 5J). CADM1 immunofluorescence staining showed that the positive cells were only present at the myenteric plexus (Figure 5K). KIT immunoreactivity and CADM1 immunoreactivity were not overlapped (Figure 5L).
Mutational Analysis of CADM1 Gene in Small Intestinal GISTs
We investigated whether small intestinal GISTs with high expression of CADM1 mRNA had the CADM1 gene mutations. cDNA sequencing analysis of CADM1 was performed in 10 duodenal/jejuno-ileal GISTs, resulted in no detection of CADM1 gene alteration (data not shown).
Discussion
CADM1 is an adhesion molecule which is implicated in tumor invasion of adult T-cell leukemia cells and acute myelocytic leukemia cells [32, 33]. Different prognoses between patients with gastric GISTs and those with small intestinal GISTs have been reported [11–13], and different protein expression between them is considered to be a possible explanation of the different prognoses. We noticed that CADM1 might be highly expressed by small intestinal GISTs compared to gastric GISTs by cDNA expression chip using a small number of gastric and small intestinal GISTs. In the present study, we tried to clarify whether higher CADM1 expression in small intestinal GISTs than gastric GISTs is really common in a large number of GIST cases, and also examined whether the expression of CADM1 is similar or not in GISTs with different types of gene abnormalities.
First, we performed CADM1 immunoblotting using eight representative gastric GISTs with exon 11 c-kit mutation and eight representative small intestinal GISTs with exon 11 c-kit mutation. All of the small intestinal GISTs showed high expression of CADM1 protein but all of the gastric GISTs did not show apparent signal. The quantitative RT-PCR also revealed that the level of CADM1 gene transcript was significantly different between gastric GISTs with exon 11 c-kit mutation and small intestinal GISTs with exon 11 c-kit mutation. We considered that gastric GISTs and small intestinal GISTs show different level of CADM1 expression even in the GISTs with the same exon 11 c-kit mutation.
In small intestinal GISTs, those with different gene abnormalities such as those with exon 9 c-kit mutation and those in NF1 patients similarly expressed high level of CADM1 mRNA as observed in those with exon 11 c-kit mutation. We concluded that CADM1 is commonly upregulated in all types of small intestinal GISTs with different gene alterations. On the other hand, most of the gastric GISTs with exon 17 c-kit mutation showed extremely low expression of CADM1 mRNA as observed in those with exon 11 c-kit mutation, but the gastric GISTs with exon 18 PDGFRA mutation expressed moderate level of CADM1 mRNA compared with those with exon 11 c-kit mutation. Thus, gastric GISTs were considered to show different level of CADM1 in those with different type of gene abnormalities. Interestingly, one gastric GIST with exon 9 c-kit mutation showed upregulation of CADM1 mRNA (data not shown). Since most GISTs with exon 9 c-kit mutation are located in the small intestine, this gastric GIST with exon 9 c-kit mutation may belong to so-called ‘small intestinal subtype of GIST’ from viewpoints of c-kit mutation type and CADM1 mRNA expression.
The different expression levels of CADM1 mRNA between gastric and small intestinal GISTs suggested that the progenitor cells (i.e. ICCs) might have different expression pattern of CADM1 in normal gastric and small intestinal walls. However, we could not detect CADM1 protein not only in gastric ICCs but also in small intestinal ICCs by double immunofluorescence staining of KIT and CADM1. Small intestinal GISTs might acquire CADM1 expression during tumorigenesis from precursor of GISTs (i.e. ICCs) to GISTs.
In our experiment, some discrepancy concerning CADM1 expression in GISTs was observed between the results of Western blotting/real-time PCR and that of immunohistochemistry. Western blotting and real-time PCR revealed that almost all small intestinal GISTs had high CADM1 expression, while immunohistochemistry showed that considerable cases of small intestinal GISTs were not apparently positive for CADM1. The antibody used might not be good enough to detect the CADM1 signal by immunohistochemistry.
As described above, expression of CADM1 might be associated with invasiveness of leukemic cells [32, 33]. Moreover, CADM1 expression in malignant pleural mesothelioma appeared to be associated with efficient adhesion and growth on CADM1 expressing mesothelium [34]. Furthermore, it has been recently reported that CADM1 might play an important role on peritoneal dissemination of tumor cells of signet ring cell carcinoma through promoting their adhesion to peritoneal cells and their growth in the serosal tissue [35]. High expression of CADM1 in small intestinal GISTs possibly resulting in promoting tumor invasion and tumor growth on the peritoneum (peritoneal dissemination) might explain the more aggressive behavior and worse prognosis of small intestinal GISTs. We are now planning to examine the role of CADM1 on invasiveness and metastasis of cultured GIST cells using in vitro and in vivo experiments. The functional experiments might clarify whether CADM1 is truly associated with worse prognosis in patients with small intestinal GISTs. Moreover, there is a possibility that targeting therapy against CADM1 has a preventive effect for vessel infiltration and peritoneal dissemination in GISTs.
Data Availability Statement
The authors acknowledge that the data presented in this study must be deposited and made publicly available in an acceptable repository, prior to publication. Frontiers cannot accept a article that does not adhere to our open data policies.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethical Committee of Hyogo College of Medicine (No. 28) and the Institutional Review Board of Osaka University Hospital (No. 08226-10). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
All of the authors participated in the collection of the clinical and pathological data. JY, TK, YH and MO carried out the expression and genetic studies. JY performed the statistical analysis. JY and SH wrote the manuscript. All authors have read the manuscript and agreed to the content.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.por-journal.com/articles/10.3389/pore.2021.602008/full#supplementary-material.
References
1. Hirota, S, Isozaki, K, Moriyama, Y, Hashimoto, K, Nishida, T, Ishiguro, S, et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science (1998) 279(5350):577–80. doi:10.1126/science.279.5350.577
2. Heinrich, MC, Corless, CL, Duensing, A, McGreevey, L, Chen, CJ, Joseph, N, et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science (2003) 299(5607):708–10. doi:10.1126/science.1079666
3. Martín-Broto, J, Rubio, L, Alemany, R, and López-Guerrero, JA. Clinical implications of KIT and PDGFRA genotyping in GIST. Clin Transl Oncol (2010) 12(10):670–6. doi:10.1007/s12094-010-0576-7
4. Antonescu, CR, Sommer, G, Sarran, L, Tschernyavsky, SJ, Riedel, E, Woodruff, JM, et al. Association of KIT exon 9 mutations with nongastric primary site and aggressive behavior: KIT mutation analysis and clinical correlates of 120 gastrointestinal stromal tumors. Clin Cancer Res (2003) 9(9):3329–37.
5. Nannini, M, Biasco, G, Astolfi, A, and Pantaleo, MA. An overview on molecular biology of KIT/PDGFRA wild type (WT) gastrointestinal stromal tumours (GIST). J Med Genet (2013) 50(10):653–61. doi:10.1136/jmedgenet-2013-101695
6. Pantaleo, MA, Astolfi, A, Astolfi, A, Urbini, M, Nannini, M, Paterini, P, et al. Analysis of all subunits, SDHA, SDHB, SDHC, SDHD, of the succinate dehydrogenase complex in KIT/PDGFRA wild-type GIST. Eur J Hum Genet (2014) 22(1):32–9. doi:10.1038/ejhg.2013.80
7. Kinoshita, K, Hirota, S, Isozaki, K, Ohashi, A, Nishida, T, Kitamura, Y, et al. Absence of c-kit gene mutations in gastrointestinal stromal tumours from neurofibromatosis type 1 patients. J Pathol (2004) 202(1):80–5. doi:10.1002/path.1487
8. Agaram, NP, Wong, GC, Guo, T, Maki, RG, Singer, S, Dematteo, RP, et al. Novel V600E BRAF mutations in imatinib-naive and imatinib-resistant gastrointestinal stromal tumors. Genes Chromosom Cancer (2008) 47(10):853–9. doi:10.1002/gcc.20589
9. Miettinen, M, and Lasota, J. Gastrointestinal stromal tumors - definition, clinical, histological, immunohistochemical, and molecular genetic features and differential diagnosis. Virchows Archiv (2001) 438(1):1–12. doi:10.1007/s004280000338
10. Joensuu, H. Risk stratification of patients diagnosed with gastrointestinal stromal tumor. Hum Pathol (2008) 39(10):1411–9. doi:10.1016/j.humpath.2008.06.025
11. Miettinen, M, and Lasota, J. Gastrointestinal stromal tumors: pathology and prognosis at different sites. Semin Diagn Pathol (2006) 23(2):70–83. doi:10.1053/j.semdp.2006.09.001
12. Emory, TS, Sobin, LH, Lukes, L, Lee, DH, and O'Leary, TJ. Prognosis of gastrointestinal smooth-muscle (stromal) tumors. Am J Surg Pathol (1999) 23(1):82–7. doi:10.1097/00000478-199901000-00009
13. Yang, Z, Wang, F, Liu, S, and Guan, W. Comparative clinical features and short-term outcomes of gastric and small intestinal gastrointestinal stromal tumours: a retrospective study. Sci Rep (2019) 9(1):10033. doi:10.1038/s41598-019-46520-1
14. Antonescu, CR, Viale, A, Sarran, L, Tschernyavsky, SJ, Gonen, M, Segal, NH, et al. Gene expression in gastrointestinal stromal tumors is distinguished by KIT genotype and anatomic site. Clin Cancer Res (2004) 10(10):3282–90. doi:10.1158/1078-0432.CCR-03-0715
15. Ichikawa, H, Yoshida, A, Kanda, T, Kosugi, Si., Ishikawa, T, Hanyu, T, et al. Prognostic significance of promyelocytic leukemia expression in gastrointestinal stromal tumor; integrated proteomic and transcriptomic analysis. Cancer Sci (2015) 106(1):115–24. doi:10.1111/cas.12565
16. Biederer, T, Sara, Y, Mozhayeva, M, Atasoy, D, Liu, XR, Kavalali, ET, et al. (2002). SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science 297 (5586):1525–31. doi:10.1126/science.1072356
17. Ito, A, and Oonuma, J. Direct interaction between nerves and mast cells mediated by the SgIGSF/SynCAM adhesion molecule. J Pharmacol Sci (2006) 102(1):1–5. doi:10.1254/jphs.cpj06014x
18. Biederer, T. Bioinformatic characterization of the SynCAM family of immunoglobulin-like domain-containing adhesion molecules. Genomics (2006) 87(1):139–50. doi:10.1016/j.ygeno.2005.08.017
19. Ito, A, Hagiyama, M, and Oonuma, J. Nerve-mast cell and smooth muscle-mast cell interaction mediated by cell adhesion molecule-1, CADM1. J Smooth Muscle Res (2008) 44(2):83–93. doi:10.1540/jsmr.44.83
20. Moiseeva, EP, Roach, KM, Leyland, ML, and Bradding, P. CADM1 is a key receptor mediating human mast cell adhesion to human lung fibroblasts and airway smooth muscle cells. PLoS One (2013) 8(4):e61579. doi:10.1371/journal.pone.0061579
21. Moiseeva, EP, Straatman, KR, Leyland, ML, and Bradding, P. CADM1 controls actin cytoskeleton assembly and regulates extracellular matrix adhesion in human mast cells. PLoS One (2014) 9(1):e85980 doi:10.1371/journal.pone.0085980
22. Wakayama, T, and Iseki, S. Role of the spermatogenic-Sertoli cell interaction through cell adhesion molecule-1 (CADM1) in spermatogenesis. Anat Sci Int (2009) 84(3):112–21. doi:10.1007/s12565-009-0034-1
23. Koma, YI, Furuno, T, Hagiyama, M, Hamaguchi, K, Nakanishi, M, Masuda, M, et al. Cell adhesion molecule 1 is a novel pancreatic-islet cell adhesion molecule that mediates nerve-islet cell interactions. Gastroenterology (2008) 134(5):1544–54. doi:10.1053/j.gastro.2008.01.081
24. Kuramochi, M, Fukuhara, H, Nobukuni, T, Kanbe, T, Maruyama, T, Ghosh, HP, et al. (2001). TSLC1 is a tumor-suppressor gene in human non-small-cell lung cancer. Nat Genet 27 (4):427–30. doi:10.1038/86934
25. Fukami, T, Fukuhara, H, Kuramochi, M, Maruyama, T, Isogai, K, Sakamoto, M, et al. Promoter methylation of theTSLC1 gene in advanced lung tumors and various cancer cell lines. Int J Cancer (2003) 107(1):53–9. doi:10.1002/ijc.11348
26. Honda, T, Tamura, G, Waki, T, Jin, Z, Sato, K, Motoyama, T, et al. Hypermethylation of theTSLC1Gene promoter in primary gastric cancers and gastric cancer cell lines. Jpn J Cancer Res (2002) 93(8):857–60. doi:10.1111/j.1349-7006.2002.tb01329.x
27. Allinen, M, Peri, L, Kujala, S, Lahti-Domenici, J, Outila, K, Karppinen, S-M, et al. Analysis of 11q21-24 loss of heterozygosity candidate target genes in breast cancer: indications of TSLC1 promoter hypermethylation. Genes Chromosom Cancer (2002) 34(4):384–9. doi:10.1002/gcc.10079
28. Takahashi, Y, Iwai, M, Kawai, T, Arakawa, A, Ito, T, Sakurai-Yageta, M, et al. Aberrant expression of tumor suppressors CADM1 and 4.1B in invasive lesions of primary breast cancer. Breast Cancer (2012) 19(3):242–52. doi:10.1007/s12282-011-0272-7
29. Lung, HL, Leung Cheung, AK, Xie, D, Cheng, Y, Kwong, FM, Murakami, Y, et al. TSLC1 is a tumor suppressor gene associated with metastasis in nasopharyngeal carcinoma. Cancer Res (2006) 66(19):9385–92. doi:10.1158/0008-5472.CAN-06-0590
30. Liu, D, Feng, X, Wu, X, Li, Z, Wang, W, Tao, Y, et al. Tumor suppressor in lung cancer 1 (TSLC1), a novel tumor suppressor gene, is implicated in the regulation of proliferation, invasion, cell cycle, apoptosis, and tumorigenicity in cutaneous squamous cell carcinoma. Tumor Biol (2013) 34(6):3773–83. doi:10.1007/s13277-013-0961-2
31. Zeng, D, Wu, X, Zheng, J, Zhuang, Y, Chen, J, Hong, C, et al. Loss of CADM1/TSLC1 expression is associated with poor clinical outcome in patients with esophageal squamous cell carcinoma. Gastroenterol Res Pract (2016) 2016:1. doi:10.1155/2016/6947623
32. Sasaki, H, Nishikata, I, Shiraga, T, Akamatsu, E, Fukami, T, Hidaka, T, et al. Overexpression of a cell adhesion molecule, TSLC1, as a possible molecular marker for acute-type adult T-cell leukemia. Blood (2005) 105(3):1204–13. doi:10.1182/blood-2004-03-1222
33. Dewan, MZ, Takamatsu, N, Hidaka, T, Hatakeyama, K, Nakahata, S, Fujisawa, J-i., et al. Critical role for TSLC1 expression in the growth and organ infiltration of adult T-cell leukemia cells in vivo. Jvi (2008) 82(23):11958–63. doi:10.1128/JVI.01149-08
34. Ito, A, Hagiyama, M, Mimura, T, Matsumoto, M, Wakayama, T, Iseki, S, et al. Expression of cell adhesion molecule 1 in malignant pleural mesothelioma as a cause of efficient adhesion and growth on mesothelium. Lab Invest (2008) 88(5):504–14. doi:10.1038/labinvest.2008.15
Keywords: gastrointestinal stromal tumor, GIST, CADM1, gastric GIST, duodenal/jejuno-ileal GIST, mutation type
Citation: Yuan J, Kihara T, Kimura N, Hashikura Y, Ohkouchi M, Isozaki K, Takahashi T, Nishida T, Ito A and Hirota S (2021) Differential Expression of CADM1 in Gastrointestinal Stromal Tumors of Different Sites and with Different Gene Abnormalities. Pathol. Oncol. Res. 27:602008. doi: 10.3389/pore.2021.602008
Received: 02 September 2020; Accepted: 25 February 2021;
Published: 19 April 2021.
Edited by:
József Timar, Semmelweis University, HungaryCopyright © 2021 Yuan, Kihara, Kimura, Hashikura, Ohkouchi, Isozaki, Takahashi, Nishida, Ito and Hirota. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seiichi Hirota, aGlyb3NAaHlvLW1lZC5hYy5qcA==
 Jiayin Yuan1
Jiayin Yuan1