- 1Clinical Genome Center, KingMed Diagnostics, Guangzhou, China
- 2Institute of KingMed Translational Medicine, Guangzhou, China
- 3Department of Pathology, KingMed Diagnostics, Guangzhou, China
Genotyping epidermal growth factor receptor (EGFR) gene in patients with advanced non-small cell lung cancers (NSCLC) is essential for identifying those patients who may benefit from targeted therapies. Systemically evaluating EGFR mutation detection rates of different methods currently used in clinical setting will provide valuable information to clinicians and laboratory scientists who take care of NSCLC patients. This study retrospectively reviewed the EGFR data obtained in our laboratory in last 10 years. A total of 21,324 NSCLC cases successfully underwent EGFR genotyping for clinical therapeutic purpose, including 5,244 cases tested by Sanger sequencing, 13,329 cases tested by real-time PCR, and 2,751 tested by next-generation sequencing (NGS). The average EGFR mutation rate was 45.1%, with 40.3% identified by Sanger sequencing, 46.5% by real-time PCR and 47.5% by NGS. Of these cases with EGFR mutations identified, 93.3% of them harbored a single EGFR mutation (92.1% with 19del or L858R, and 7.9% with uncommon mutations) and 6.7% harbored complex EGFR mutations. Of the 72 distinct EGFR variants identified in this study, 15 of them (single or complex EGFR mutations) were newly identified in NSCLC. For these cases with EGFR mutations tested by NGS, 65.3% of them also carried tumor-related variants in some non-EGFR genes and about one third of them were considered candidates of targeted drugs. NGS method showed advantages over Sanger sequencing and real-time PCR not only by providing the highest mutation detection rate of EGFR but also by identifying actionable non-EGFR mutations with targeted drugs in clinical setting.
Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide [1, 2]. Approximately 610,000 lung cancer-related deaths were reported in China in 2015 [3]. Non-small cell lung cancer (NSCLC) is the most common histological subtype of lung cancer, accounting for approximately 80–85% of the disease. Targeted therapy based on the identification of actionable genetic/genomic alterations in the disease has led to integration of molecular testing for planning treatment strategies for advanced NSCLC patients [4, 5].
Activating mutation in the epidermal growth factor receptor gene (EGFR) is the most frequent genetic alteration in NSCLC. EGFR mutations can be detected in 15% of adenocarcinoma subtype of Caucasian NSCLC but in 40–50% of same subtype of East Asian NSCLC. Exon-19 deletions (19del) and L858R substitution in exon-21 are the 2 classical EGFR mutations which could predict tumor responses to EGFR tyrosine kinase inhibitors (EGFR-TKIs) in NSCLC patients [6–10]. Other uncommon EGFR mutations have also been found to show sensitiveness (eg., Exon-19 insertions, p.L861Q in exon-21, p.G719X in exon-18, and p.S768I in exon-20) or resistance (eg., most exon-20 insertions) to EGFR-TKIs. T790M substitution in exon-20 is a well-known acquired mutation resistant to first or second generation EGFR-TKIs, but sensitive to third generation EGFR-TKIs. Currently, genotyping EGFR has been recommended by both laboratory and clinical guidelines as evidence-based standard care for advanced NSCLC patients [11–14].
Various technical platforms are clinically available for genotyping EGFR, including commonly used methods, such as Sanger sequencing, real-time PCR and NGS and occasionally used methods, such as denaturing high performance liquid chromatography (DHPLC) and Luminex liquid chip. Systemically comparing EGFR mutation detection rates detected by these methods routinely used in clinical setting is rarely reported [15, 16]. In this study, we retrospectively reviewed our successfully tested EGFR results for 21,324 unselected Chinese NSCLC patients whose specimens were performed in our laboratory, a College of American Pathologists (CAP)-certified reference laboratory providing EGFR mutation testing for NSCLC patients.
Materials and Methods
Patients
In total, 21,324 Formalin-Fixed and Paraffin-Embedded (FFPE) tumor specimens of NSCLC patients from 30 provinces of China were successfully performed in our laboratory for testing EGFR mutations from June 2009 to December 2018. The EGFR testing results were retrospectively reviewed and reported with exclusion of those cases with failed EGFR testing. For the cases with duplicated tests, only the result of the first successful test was counted. Ages of these patients ranged from 16 to 96 years old (median: 63 years old) and the gender percentages were 56.9% for females, 41.5% for males, and 1.6% with unknown gender information. All the EGFR tests were ordered by physicians for therapeutic purposes and were performed at a single testing center. This study was approved by the ethics review board of KingMed Diagnostics.
Identification of Epidermal Growth Factor Receptor Mutations
The numbers of cases tested by different methods were shown in Supplementary Figure S1. According to the standard operation procedures (SOPs) validated in our laboratory, prior to EGFR mutation testing, tumor cell content (TCC) of the specimens was assessed by at least an experienced pathologist. A specimen with ≥20% TCC was required by Sanger sequencing, and recommended for real-time PCR and NGS methods. A specimen with 1–20% TCC was acceptable for testing via real-time PCR or NGS but not by Sanger sequencing. DNA from the specimens was extracted using QIAamp DNA FFPE Tissue Kit according to the manufacturer’s protocol (Qiagen China, Shanghai, China). MagMAX FFPE DNA/RNA Ultra Kit was used to isolate both DNA and RNA from the same section of FFPE tissues for the amplicon-based NGS testing (Life Technologies Corporation, Austin, United States). Technical parameters, such as sensitivities and specificities etc., were determined before these methods were used in clinical setting (data not shown).
Sanger sequencing for genotyping EGFR was launched since the year of 2009 and a total of 5,244 cases successfully tested by this technique was included in this study. EGFR exons-18 to -21 were amplified using polymerase chain reaction (PCR) and directly sequenced using ABI 3730xl (Applied Biosystems, Foster City, United States).
Since 2013, real-time PCR-based method for detecting EGFR mutations was applied in our laboratory using a commercial kit, EGFR RGQ PCR Kit (Qiagen China, Shanghai, China). A total of 13,329 cases successfully tested by this technique was included in this study. This technique covers 29 known mutations spanning exons-18 to -21 including exon-18 missense mutations at G719X (G719S, G719A and G719C), exon-19 deletions (19del), exon-20 missense mutations (S768I and T790M) and insertions (20ins), and exon-21 missense mutations (L858R and L861Q). Genotyping EGFR using NGS was launched in 2016 and so far, a total of 2,751 cases was successfully performed and the results were included in this study. For the first 1,089 cases, Ion AmpliSeq™ Colon and Lung Cancer Panel V2 and Ion AmpliSeq™ RNA Fusion panel were used covering point mutations and small insertions and deletions (indels) of 22 genes (EGFR, ALK, BRAF, KRAS, MET, ERBB2, AKT1, CTNNB1, ERBB4, DDR2, FBXW7, FGFR1, FGFR2, FGFR3, MAP2K1, NOTCH1, NRAS, PIK3CA, PTEN, SMAD4, STK11 and TP53) as well as fusions of 4 genes (ALK, ROS1, RET and NTRK1) (ThermoFisher, Waltham, United States). For these cases, sequencing was performed on an Ion Torrent PGM instrument and data analysis was performed using Torrent Suite Software and Torrent Server. For the remaining 1,662 cases, we adopted a validated capture-based method for library preparation (Integrated DNA Technologies, Inc., Coralville, United States) and performed DNA sequencing using Illumina Nextseq 500 or NovaSeq 6000 systems (Illumina, San Diego, United States). After sequencing, a clinically validated bioinformatics pipeline was used to identify variants in the targeted genes. Sequence variants were interpreted and reported according to the guideline compiled by Association of Molecular Pathology (AMP) [17]. In brief, tier 1 (variants of strong clinical significance) and tier 2 (variants of potential clinical significance) variants were reported while tier 3 (variants of unknown clinical significance) and tier 4 (benign or likely benign variants) variants were not reported. Detailed technical procedures for all the 3 methods were listed in Supplementary Material.
Statistical Analysis
We used Chi-square testing for comparing EGFR mutation detection rates identified by the 3 methods. p-values less than 0.05 were considered statistically significant in all scenarios. IBM SPSS Statistics Version 19 was used for all statistical analysis.
Results
Tumor Cell Content in Different Testing Groups
Our records showed that 5,244 NSCLC samples with 20–90% TCCwere successfully tested by Sanger sequencing, 13,329 NSCLC samples with 1–95% TCC were tested by real-time PCR, and 2,751 NSCLC samples with 1–90% TCC were tested by NGS. In details, 58.5% (12,484/21,324) of the samples were recorded with <40% TCC accounting for 46.8% (2,454/5,244) of the samples tested by Sanger sequencing, 60.8% (8,109/13,329) tested by real-time PCR, and 69.8% (1,921/2,751) tested by NGS; 20.4% (4,359/21,324) of the samples were recorded with <20% TCC accounting for 26.3% (3,509/13,329) tested by real-time PCR and 30.9% (850/2,751) tested by NGS; 2.3% (490/21,324) of the samples were recorded with 1% TCC accounting for 3.2% (429/13,329) tested by real-time PCR and 2.2% (61/2,751) tested by NGS (Table 1).

TABLE 1. Percentages of NSCLC specimens with different tumor cell content (TCC) tested by different platforms.
Epidermal Growth Factor Receptor Mutation Rates
Of the 21,324 NSCLC samples successfully tested for EGFR, 9,621 of them carried somatic EGFR mutations, representing an average positive EGFR detection rate of 45.1% including 40.3% (2,111/5,244) tested by Sanger sequencing, 46.5% (6,202/13,329) tested by real-time PCR, and 47.5% (1,308/2,751) tested by NGS respectively (Figure 1A.). The detection rates obtained by real-time PCR and NGS methods were significantly higher than that obtained by Sanger sequencing (p < 0.001) (Figure 1A).
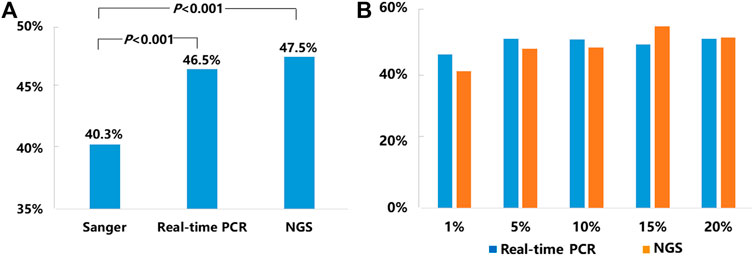
FIGURE 1. Mutation detection rates for EGFR. (A) The average mutation detection rates for EGFR obtained by Sanger sequencing, real-time PCR, and NGS methods. (B)EGFR mutation detection rates in NSCLC samples with 1–20% TCC by real-time PCR and NGS.
Since our SOPs allowed testing NSCLC samples with TCC from 1 to 20% by both real-time PCR and NGS methods, we compared EGFR mutation detection rates by these two methods in the samples with different levels of TCC (1, 5, 10, 15, and 20%). Detailed information was listed in Figure 1B. In brief, the positive detection rates of real-time PCR vs. NGS were 45.9% (197/429) vs. 41% (25/61) in 1% TCC, 50.7% (380/750) vs. 47.6% (79/166) in 5% TCC, 50.4% (765/1,518) vs. 48% (195/406) in 10% TCC, 49% (398/812) vs. 54.4% (118/217) in 15% TCC, and 50.6% (1,077/2,129) vs. 51.1% (258/505) in 20% TCC. Statistically, there were no significant difference about the detection rates by the 2 methods in each of the 5 subgroups (p > 0.05), although real-time PCR method showed higher detection rates in relatively lower TCC (1, 5, and 10%) in contrast to the higher detection rates in samples with relatively higher TCC (15 and 20%) by NGS.
Since NGS is a quantitative method for EGFR testing, we analyzed EGFR variant allele frequencies (VAFs) in the specimens with different TCC levels. The median EGFR VAFs were 0.074 (range: 0.01–0.675), 0.097 (range: 0.011–0.97), 0.161 (range: 0.01–0.902), 0.254 (range: 0.014–0.811), and 0.261 (range: 0.01–0.957) for NSCLC samples with 1, 5, 10, 15, and ≥20% TCC respectively. Although the median EGFR VAFs increase with increased TCC levels in the specimens tested, the values of EGFR VAFs overlapped greatly among groups with different TCC, showing very high EGFR VAF variations ranging from 0.01 to 0.97 (for example, EGFR VAF was up to 0.675 in one sample with 1% TCC, down to 0.01 in another sample with 20% TCC).
Epidermal Growth Factor Receptor Mutation Patterns
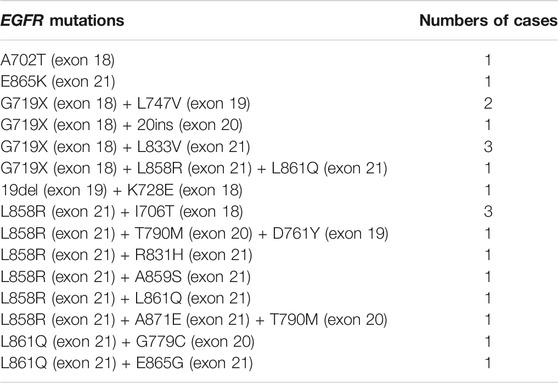
Of the 9,621 cases with EGFR mutations, 93.3% of them (8,979/9,621) harbored a single EGFR mutation, including 8,274 (92.1%) cases carrying classical mutations (19del or L858R) and 705 (7.9%) cases carrying rare mutations while 6.7% of them (642/9,621) harbored complex EGFR mutations. Distribution of these EGFR mutations were shown in Figures 2A,B. The majority of the complex EGFR mutations occurred with coexistence of T790M (exon-20), G719X (exon-18), L858R (exon-21) and 19del (Figure 2B).
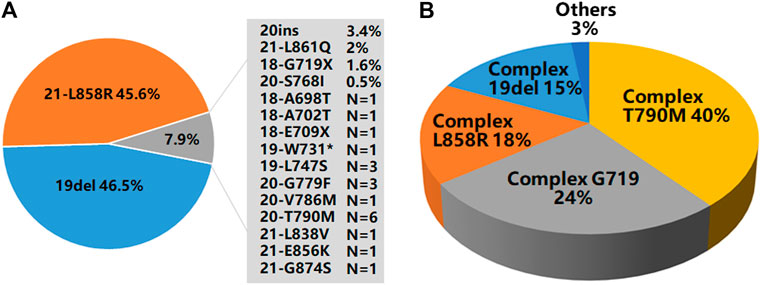
FIGURE 2. EGFR mutation patterns. (A) An overview of EGFR mutations present in 8,979 cases with single EGFR mutations. (B) A summary of EGFR mutations present in 642 cases with complex EGFR mutations.
Among the EGFR mutations identified in the 9,621 cases, a total of 72 distinct variants could be recognized (different exon-19 deletions were classified as 19del, various exon-20 insertions were grouped as 20ins, and different G719, E709 and R776 mutations were labeled as G719X, E709X and R776X). NGS, Sanger sequencing and real-time PCR detected 76, 46, and 36% of the distinct EGFR mutations respectively. For the cases with single uncommon EGFR mutation, NGS detected 94% of them, while Sanger sequencing and real-time PCR detected 41% of them. For complex EGFR mutations, NGS detected 71% of them, while Sanger sequencing and real-time PCR detected 47% and 35% respectively. Details about these EGFR mutations were shown in Supplementary Tables S1 and S2. Of the 72 distinct EGFR variants, 15 of them (single or complex EGFR mutations) were newly identified in NSCLC (11 of them detected by NGS, 2 of them detected by Sanger sequencing and 3 detected by real-time PCR, with 1 detected by both Sanger sequencing and NGS). These newly identified variants were shown in Table 2.
Actionable Mutations in Non-Epidermal Growth Factor Receptor Genes Identified by Next-Generation Sequencing
Of the 2,751 cases tested by NGS, 52.5% of them (1,443/2,751) were found to be EGFR mutation negative. The 1,443 cases could be grouped into 3 subgroups: 1) 27% (N = 742,742/2,751) of them were found to have actionable mutations in the non-EGFR genes with available targeted drugs or potential drugs under clinical trials, including ALK, ROS1, RET fusions and other oncogenic events (Figure 3A) as well as MAP2K1 (0.1%), AKT1 (0.1%) and FGFR3 (0.1%) (Data not shown in Figure 3A); 2) 18.3% (N = 504,504/2,751) were found to have tumor initiation- and/or progression-related mutations in non-EGFR genes, such as TP53, SMAD4, FBXW7, CTNNB1 and NOTCH1. However there are no targeted drugs or potential drugs available to these mutated genes products currently; 3) 7.2% (N = 197,197/2,751) of them were found to carry neither oncogenic mutations in EGFR nor tumor initiation- and/or progression-related mutations in non-EGFR genes.
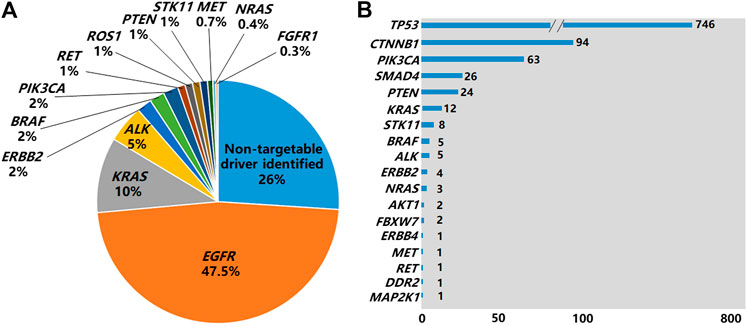
FIGURE 3. Non-EGFR genes carrying mutations identified by NGS. (A) Genes carrying targetable driver mutations identified in the 2,751 NSCLC cases by NGS. (B) Non-EGFR genes carrying mutations identified by NGS in 1,308 EGFR-positive cases by NGS.
Of the 1,308 cases with EGFR mutations found by NGS, 65.3% of the cases (854/1,308) harbored non-EGFR mutations in 18 tumor-related genes, with TP53 being the most frequently mutated (Figure 3B). The overall co-existing frequencies of EGFR/KRAS, EGFR/ALK, EGFR/BRAF, EGFR/NRAS were 0.4% (12/2,751), 0.2% (5/2,751), 0.2% (5/2,751) and 0.1% (3/2,751) respectively. EGFR/KRAS co-mutations were found in 0.9% (12/1,308) of the 1,308 patients with EGFR mutations and 4% (12/299) of the 299 patients with KRAS mutations. EGFR/ALK co-mutations were found in 3.8% (5/131) of the 131 patients with ALK fusions. EGFR/BRAF co-mutations were detected in 10.6% (5/47) of the 47 patients with BRAF mutations. EGFR/NRAS co-mutations were detected in 30% (3/10) of the 10 patients with NRAS mutations. In addition, we found 1 patient carrying both EGFR mutation and RET fusion. No patient was found to carry EGFR mutation co-existing with ROS1 fusion or Met exon 14 skipping mutation in this cohort (Supplementary Table S4). Altogether, 83.8% (716/854) of patients carried mutations in 2 genes, 14.6% (125/854) of them carried mutations in 3 genes, and 1.5% (13/854) of them carried mutations in 4 genes (Supplementary Table S3).
We compared the mutation rates of some driver genes including EGFR, ALK, ROS1, RET and KRAS by the two different platforms of NGS testing. The positive detection rates of amplicon-based vs capture-based sequencing were 47.8% (521/1089) vs. 47.4% (787/1662) for EGFR, 5.7% (62/1089) vs. 4.2% (69/1662) for ALK fusions, 0.9% (10/1089) vs. 1% (17/1662) for ROS1 fusions, 1.3% (14/1089) vs. 1.5% (25/1662) for RET fusions. Statistically, there were no significant difference about the detection rates for these driver gene mutations by the 2 NGS methods (p > 0.05), although amplicon-based method showed relatively higher detection rates for ALK fusions compared to capture-based method.
Discussion
To our knowledge, this research project represents the largest data analysis of EGFR mutational status in Chinese patients with NSCLC by multiple platforms, providing several interesting findings valuable in clinical settings.
NGS testing expanded mutational spectrum of EGFR in NSCLC patients. Although only 2,751 of the 21,324 (12.9%) NSCLC specimens were tested by NGS, we identified 11 novel EGFR mutations (not being previously reported in NSCLC) in exons-18 to -21 by the NGS methods while Sanger sequencing and real-time PCR identified only 2 and 3 novel EGFR mutations in this region respectively (Supplementary Tables S1 and S2). We believe that if all of the 21,324 NSCLC specimens had been analyzed by NGS, all of the rare variants found by Sanger sequencing and real-time PCR would have been identified. Although both NGS and Sanger sequencing detected the region covering exons-18 to -21 of EGFR, Sanger sequencing will miss those EGFR mutations with frequencies of mutant alleles less than 20% (cut-off value determined in our validated data) due to its low technical sensitivity. Both NGS and real-time PCR showed similar technical sensitivity (1% frequency of mutant alleles), but real-time PCR method was designed to detect only 29 hotspot mutations of EGFR and might have missed mutations in the non-hotspot regions. Furthermore, real-time PCR couldn’t distinguish the differences within or around 19del, G719X, 20ins variants, for example G719A or G719C, though these variants may show different responses to EGFR-TKIs [18–20] (Table 3). In summary, NGS method shows obvious advantages over Sanger sequencing and real-time PCR methods for identifying novel actionable EGFR mutations.

TABLE 3. A summary of EGFR mutations tested by Sanger sequencing, real-time PCR and NGS platforms for NSCLC patients in this project.
NGS identified a long list of non-EGFR mutations related to tumor initiation and progression, adding additional therapeutic opportunities and/or assessing the prognostic outcomes for NSCLC patients. In this study, 52.5% (1,443/2,751) of the cases tested by NGS did not carry EGFR mutations, similar to the results reported in previous Asian lung adenocarcinoma series [10]. Of the cases tested by NGS, 27% of them were found to have actionable mutations, including ALK (5%), ROS1 (1%), BRAF (2%), KRAS (10%), RET (1%), ERBB2 (HER2) (2%), and exon-14 skipping mutations of MET (0.7%) as well as other genes, such as PIK3CA (2%), PTEN (1%), STK11 (1%) NRAS (<0.5%), FGFR1 (<0.5%), FGFR3 (<0.5%), MAP2K1 (<0.5%), and AKT1 (<0.5%). For ALK, ROS1 and BRAF V600E alterations, targeted drugs have been available in clinical setting. For the other genes in this list, potential targeted drugs providing possible therapeutic opportunities for NSCLC patients are emerging in some international clinical trials (www.clinicaltrials.gov). In addition, 18.3% of the cases tested by NGS carried tumor-mutations in non-EGFR genes classified currently as non-actionable, such as TP53, SMAD4, FBXW7, CTNNB1 and NOTCH1. However, pathogenic variants in these genes were considered to have prognostic or predictive significances for NSCLC patients [21, 22]. For the 7.2% of the cases tested by NGS without finding any of tier 1 and tier 2 mutations in neither EGFR nor non-EGFR genes, but we think tier 3 variants (variants of unknown clinical significance-VUS, not reported) found in these cases might represent potentially useful biomarkers for monitoring cancer treatment effects via liquid biopsy. Interestingly, of the 1,308 cases with EGFR mutations found by NGS, 65.3% of the cases (854/1,308) also harbored non-EGFR mutations in 18 tumor-related genes. In NSCLC, oncogenic driver mutations are typically mutually exclusive. However, cases of mutations in multiple driver genes are increasingly reported, as well as in our retrospective study. EGFR and other driver genes including KRAS, ALK, BRAF, NRAS, RET co-alterations are likely to represent certain proportion of cases with multiple mutations in NSCLC. These co-alterations may provide prognostic or predictive effects to EGFR-TKIs as reported previously [23–26].
NGS is the method which could maximize the findings of EGFR mutations as shown in this project. The total EGFR positive mutation rate in our data was 45.1%, concordant with several previous studies of Chinese patients with NSCLC [10, 16]. By comparing the EGFR mutation detection rates showed by the 3 methods, 40.3% (2,111/5,244) by Sanger sequencing was significantly lower than that found by real-time PCR (46.5%, 6,202/13,329) and NGS (47.5%, 1,308/2,751) (p < 0.001), indicating that the Sanger sequencing had missed some EGFR mutations which would have been identified if NGS or real-time PCR methods had been applied. The main reason leading to higher detection rates by NGS or real-time PCR is attributed to higher sensitivities of the NGS and real-time PCR (1% frequency of mutant alleles) than that of Sanger sequencing (20% frequency of mutant alleles) (Table 3). As expected, comparable EGFR-positive rates identified by real-time PCR and NGS were observed in the current study. By stratifying different levels of TCC from 1 to 20% in tumor specimens for analyzing EGFR detection rates, there were no significant differences for EGFR positive mutations rates by either real-time PCR or NGS. This finding suggested that EGFR mutations present in these specimens with TCC from 1 to 20% could be fairly identified by real-time PCR or NGS, providing therapeutic opportunities using EGFR-TKIs for these patients. EGFR mutation rates by Sanger sequencing in the FFPE samples with <40 and ≥40% TCC were 30.8% (757/2,454) and 48.5% (1,354/2790) respectively, showing similar EGFR positive rates in FFPE samples with TCC ≥40% by the 3 methods. Based on these findings, we recommend EGFR mutation testing of specimens with ≥1% TCC by NGS or real-time PCR. Sanger sequencing could be considered for specimens with ≥40% TCC due to its low cost (Table 3). However, considering the facts that the EGFR VAFs overlapped greatly among groups with different TCC, showing high levels of variations regarding EGFR VAFs ranging from 0.01 to 0.97 (EGFR VAF was up to 0.675 in one sample with 1% TCC, down to 0.01 in another sample with 20% TCC), some low level of EGFR VAFs in high level TCC specimens could have been missed by Sanger sequencing method.
We acknowledge that there are several limitations or concerns in this research: 1) Since some features, such as pathologic diagnosis (adenocarcinoma, squamous or other histological types), smoking status, grades, and stages of the disease, were not fully described in their requisition forms when EGFR testing were ordered for therapeutic purpose, further stratification analysis were not implemented. 2) Follow-up data were not available for monitoring responses of TKIs treatment targeting to those uncommon EGFR mutations, complex EGFR mutations or co-existing mutations in both EGFR and non-EGFR genes. Even for NSCLC patients with a classical EGFR mutation (L858R in exon-21 or 19del), it was speculated that different outcomes after EGFR-TKIs treatments might be present [27]. 3) Although some identified novel variants of EGFR and non-EGFR were considered relevant to the initiation and progression of NSCLC, further investigations about their abnormal functions and pathogenicity are required.
In conclusions, we presented a large dataset of EGFR mutations in Chinese NSCLC patients including a long list of novel EGFR mutations identified, providing valuable information for the diagnosis and subsequent treatment of the disease. NGS method showed advantages over Sanger sequencing and real-time PCR not only by providing the highest mutation detection rates of EGFR but also by identifying actionable non-EGFR mutations with available targeted drugs in clinical setting.
Data Availability Statement
Data are available from the corresponding author on reasonable request.
Ethics Statement
This study was approved by the ethics review board of KingMed Diagnostics.
Author Contributions
LM and SY conceptualized the study; XHO, YX, YT, XYO and CH performed experiments; WZ, XL, SZ, CZ, DZ, XD and PL performed the data analysis; LM and SY wrote the manuscript with input from all authors; All authors have read and approved the final version of the manuscript.
Funding
The authors declare that this study received funding from Science and Technology Program of Guangzhou, China (201802020030), Program for Entrepreneurial and Innovative Leading Talents of Guangzhou, China (CXLJTD-201603), Science and Technology Program of Guangzhou, China (201604046001), Project of Academician Workstation at KingMed Diagnostics (2017B090904030), Program for Entrepreneurial and Innovative Leading Talents of Guangzhou, China (201809010012). The funder had the following involvement with the study: Mutation Spectrum of EGFR from 21,324 Chinese Patients with Non-Small Cell Lung Cancer (NSCLC) Successfully Tested by Multiple Methods in a CAP-Accredited Laboratory. This study did not receive any commercial funding.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to all the patients for their medical information included in this project.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.por-journal.com/articles/10.3389/pore.2021.602726/full#supplementary-material.
References
1. Bray, F, Ferlay, J, Soerjomataram, I, Siegel, RL, Torre, LA, and Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2018) 68:394–424. doi:10.3322/caac.21492
2. Siegel, RL, Miller, KD, and Jemal, A. Cancer statistics, 2019. CA A Cancer J Clin (2019) 69:7–34. doi:10.3322/caac.21551
3. Chen, W, Zheng, R, Baade, PD, Zhang, S, Zeng, H, Bray, F, et al. Cancer statistics in China, 2015. CA Cancer J Clin (2016) 66:115–32. doi:10.3322/caac.21338
4. Gridelli, C, Rossi, A, Carbone, DP, Guarize, J, Karachaliou, N, Mok, T, et al. Non-small-cell lung cancer. Nat Rev Dis Primers (2015) 1:15009. doi:10.1038/nrdp.2015.9
5. Kumarakulasinghe, NB, van Zanwijk, N, and Soo, RA. Molecular targeted therapy in the treatment of advanced stage non-small cell lung cancer (NSCLC). Respirology (2015) 20:370–8. doi:10.1111/resp.12490
6. Mok, TS, Wu, YL, Thongprasert, S, Yang, CH, Chu, DT, Saijo, N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med (2009) 361:947–57. doi:10.1056/nejmoa0810699
7. Mitsudomi, T, Morita, S, Yatabe, Y, Negoro, S, Okamoto, I, Tsurutani, J, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol (2010) 11:121–8. doi:10.1016/s1470-2045(09)70364-x
8. Zhou, C, Wu, YL, Chen, G, Feng, J, Liu, XQ, Wang, C, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol (2010) 12:735–42. doi:10.1016/S1470-2045(11)70184-X
9. Castellanos, E, Feld, E, and Horn, L Driven by mutations: the predictive value of mutation subtype in EGFR -mutated non-small cell lung cancer. J Thorac Oncol (2017) 12:612–23. doi:10.1016/j.jtho.2016.12.014
10. Dearden, S, Stevens, J, Wu, YL, and Blowers, D. Mutation incidence and coincidence in non small-cell lung cancer: meta-analyses by ethnicity and histology (mutMap). Ann Oncol (2013) 24:2371–6. doi:10.1093/annonc/mdt205
11.National Comprehensive Cancer Network. NCCN clinical practice guidelines in Oncology (NCCN Guidelines®) non-small cell lung cancer (2020). Available from: https://www.nccn.org/professionals/physician_gls/default.aspx#nscl/ (Accessed 7 August 2020).
12. Planchard, D, Popat, S, Kerr, K, Novello, S, Smit, EF, Faivre-Finn, C, et al. Correction to: “metastatic non-small cell lung cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up”. Ann Oncol (2019) 30:863–70. doi:10.1093/annonc/mdy474
13.National Health Commission of the People's Republic of China. Chinese guidelines for diagnosis and treatment of primary lung cancer 2018 (English version). Chin J Cancer Res (2019) 31:1–28. doi:10.21147/j.issn.1000-9604.2019.01.01
14. Kalemkerian, GP, Narula, N, Kennedy, EB, Biermann, WA, Donington, J, Leighl, NB, et al. Molecular testing guideline for the selection of patients with lung cancer for treatment with targeted tyrosine kinase inhibitors: American society of clinical oncology endorsement of the college of American pathologists/international association for the study of lung cancer/association for molecular pathology clinical practice guideline update. J Clin Oncol (2018) 36:911–9. doi:10.1200/jco.2017.76.7293
15. Bai, H, Wang, Z, Wang, Y, Zhuo, M, Zhou, Q, Duan, J, et al. Detection and clinical significance of intratumoral EGFR mutational heterogeneity in Chinese patients with advanced non-small cell lung cancer. PLoS One (2013) 8:e54170. doi:10.1371/journal.pone.0054170
16. Cheng, Y, Wang, Y, Zhao, J, Liu, Y, Gao, H, Ma, K, et al. Real-world EGFR testing in patients with stage IIIB/IV non-small-cell lung cancer in North China: a multicenter, non-interventional study. Thorac Cancer (2018) 9:1461–9. doi:10.1111/1759-7714.12859
17. Li, MM, Datto, M, Duncavage, EJ, Kulkarni, S, Lindeman, NI, Roy, S, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer. J Mol Diagn (2017) 19:4–23. doi:10.1016/j.jmoldx.2016.10.002
18. Wu, JY, Yu, CJ, Chang, YC, Yang, CH, Shih, JY, and Yang, PC. Effectiveness of tyrosine kinase inhibitors on “uncommon” epidermal growth factor receptor mutations of unknown clinical significance in non-small cell lung cancer. Clin Cancer Res (2011) 17:3812–21. doi:10.1158/1078-0432.ccr-10-3408
19. Tu, HY, Ke, EE, Yang, JJ, Sun, YL, Yan, HH, Zheng, MY, et al. A comprehensive review of uncommon EGFR mutations in patients with non-small cell lung cancer. Lung Cancer (2017) 114:96–102. doi:10.1016/j.lungcan.2017.11.005
20. Massarelli, E, Johnson, FM, Erickson, HS, Wistuba, II, and Papadimitrakopoulou, V Uncommon epidermal growth factor receptor mutations in non-small cell lung cancer and their mechanisms of EGFR tyrosine kinase inhibitors sensitivity and resistance. Lung Cancer (2013) 80:235–41. doi:10.1016/j.lungcan.2013.01.018
21. Liu, Y, Xu, F, Wang, Y, Wu, Q, Wang, B, Yao, Y, et al. Mutations in exon 8 of TP53 are associated with shorter survival in patients with advanced lung cancer. Oncol Lett (2019) 18:3159–69. doi:10.3892/ol.2019.10625
22. Xiao, Y, Yin, C, Wang, Y, Lv, H, Wang, W, Huang, Y, et al. FBXW 7 deletion contributes to lung tumor development and confers resistance to gefitinib therapy. Mol Oncol (2018) 12:883–95. doi:10.1002/1878-0261.12200
23. Labbé, C, Cabanero, M, Korpanty, GJ, Tomasini, P, Doherty, MK, Mascaux, C., et al. Prognostic and predictive effects of TP53 co-mutation in patients with EGFR -mutated non-small cell lung cancer (NSCLC). Lung Cancer (2017) 111:23–9. doi:10.1016/j.lungcan.2017.06.014
24. Skoulidis, F, and Heymach, JV Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer (2019) 19:495–509. doi:10.1038/s41568-019-0179-8
25. Tsukumo, Y, Naito, M, and Suzuki, T Influence of EGFR-activating mutations on sensitivity to tyrosine kinase inhibitors in a KRAS mutant non-small cell lung cancer cell line. PLoS One (2020) 15:e0229712. doi:10.1371/journal.pone.0229712
26. Yang, J-J, Zhang, XC, Su, J, Xu, CR, Zhou, Q, Tian, HX, et al. Lung cancers with concomitant EGFR mutations and ALK rearrangements: diverse responses to EGFR-TKI and crizotinib in relation to diverse receptors phosphorylation. Clin Cancer Res (2014) 20:1383–92. doi:10.1158/1078-0432.ccr-13-0699
27. Lee, CK, Wu, Y-L, Ding, PN, Lord, SJ, Inoue, A, Zhou, C, et al. Impact of specific epidermal growth factor receptor (EGFR) mutations and clinical characteristics on outcomes after treatment with EGFR tyrosine kinase inhibitors versus chemotherapy in EGFR-mutant lung cancer: a meta-analysis. J Clin Oncol (2015) 33:1958–65. doi:10.1200/jco.2014.58.1736
Keywords: non-small cell lung cancer, EGFR mutation testing, Sanger sequencing, real-time PCR, next-generation sequencing
Citation: Mao L, Zhao W, Li X, Zhang S, Zhou C, Zhou D, Ou X, Xu Y, Tang Y, Ou X, Hu C, Ding X, Luo P and Yu S (2021) Mutation Spectrum of EGFR From 21,324 Chinese Patients With Non-Small Cell Lung Cancer (NSCLC) Successfully Tested by Multiple Methods in a CAP-Accredited Laboratory. Pathol. Oncol. Res. 27:602726. doi: 10.3389/pore.2021.602726
Received: 04 September 2020; Accepted: 01 March 2021;
Published: 07 April 2021.
Edited by:
József Tímár, Semmelweis University, HungaryCopyright © 2021 Mao, Zhao, Li, Zhang, Zhou, Zhou, Ou, Xu, Tang, Ou, Hu, Ding, Luo and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shihui Yu, emIteXVzaGlodWlAa2luZ21lZC5jb20uY24=
†These authors have contributed equally to this work
 Linlin Mao1†
Linlin Mao1†