In the original article, there were mistakes in the legend for all of the figures as published. The source of the pictures cited in the article were not included. The correct legends appear below.
FIGURE 1
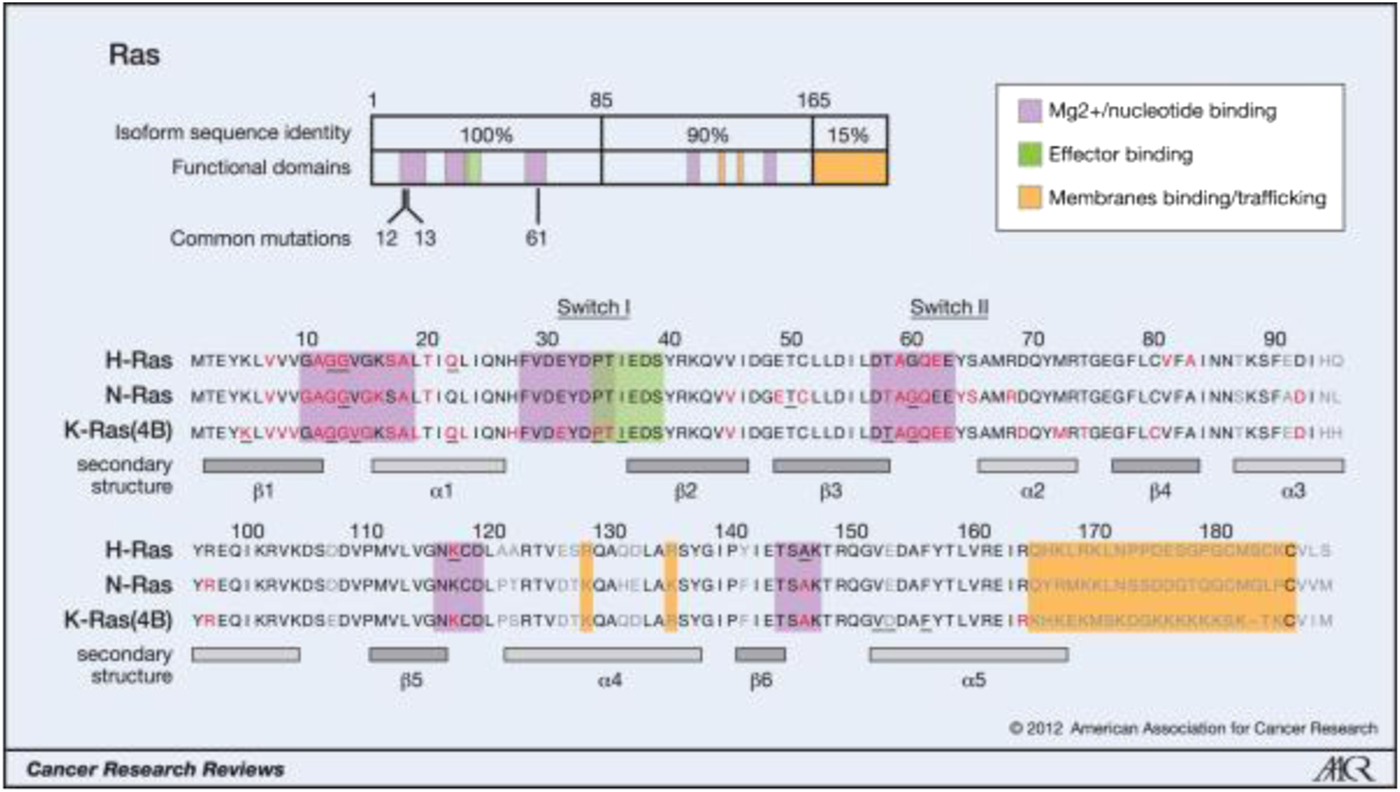
Oncogenic mutations of Ras isoforms. (Reprinted by permission from Cancer Research [3]).
FIGURE 2
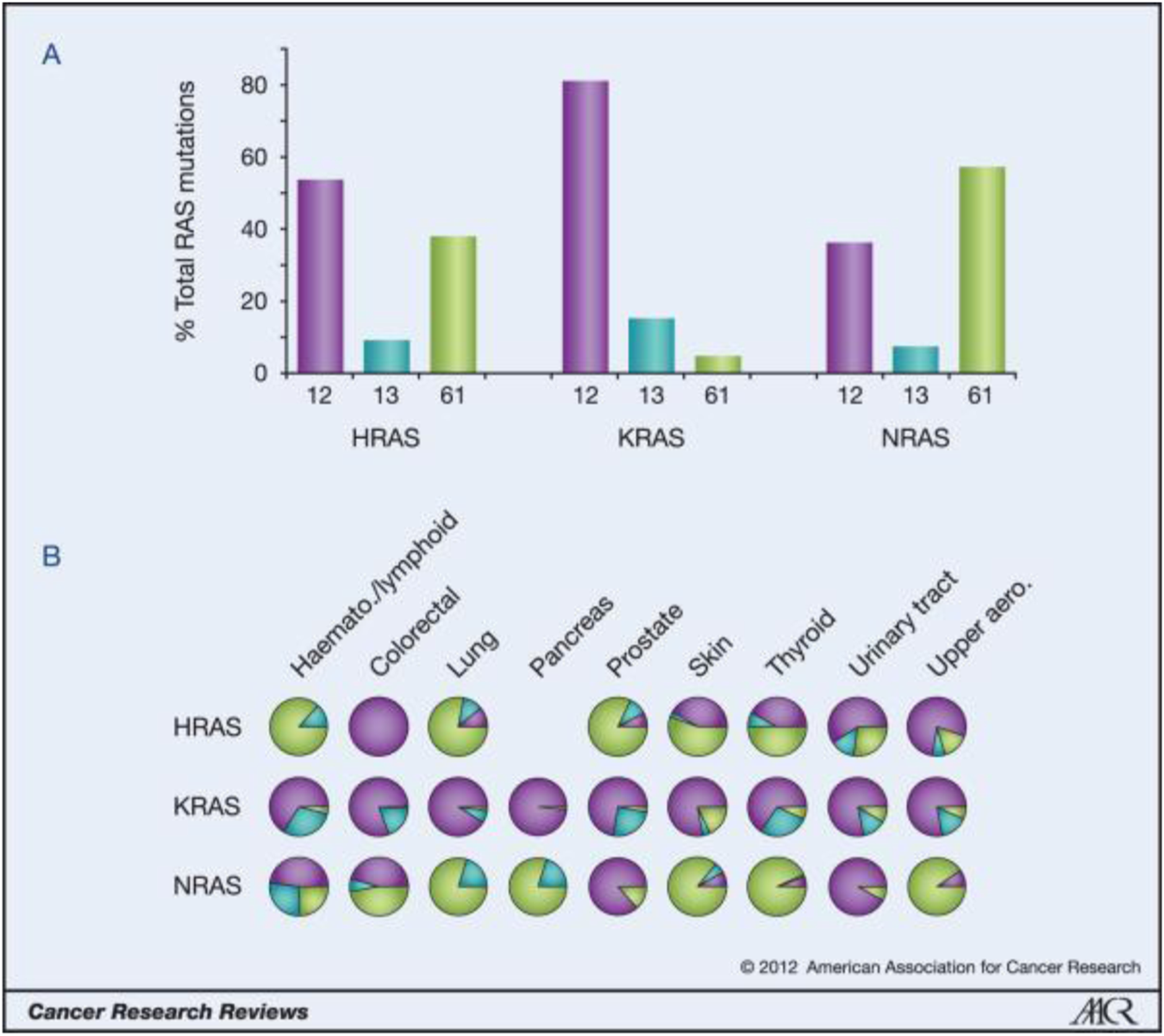
Ras isoform-specific codon mutation bias. (Reprinted by permission from Cancer Research [3]).
FIGURE 3
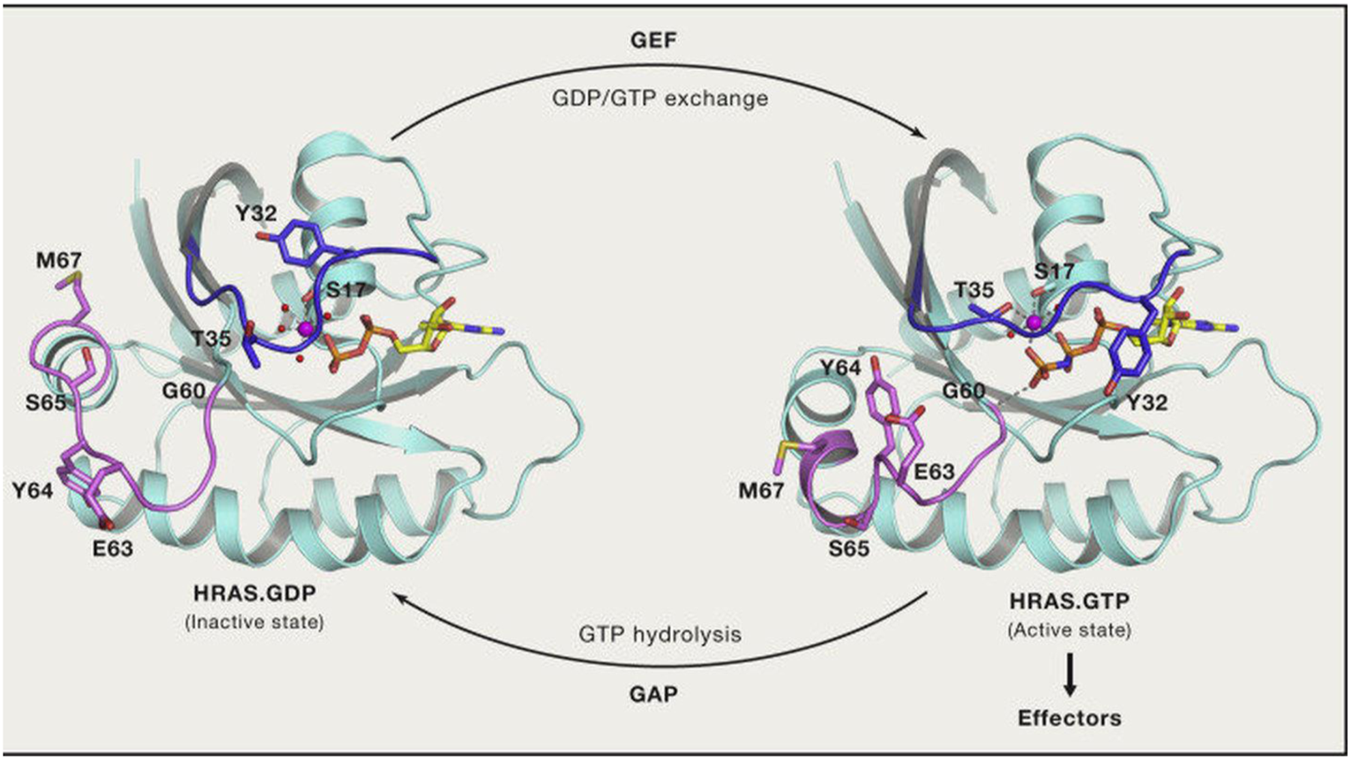
RAS GTP/GDP change. (Reprinted by permission from Cell [4]).
FIGURE 4
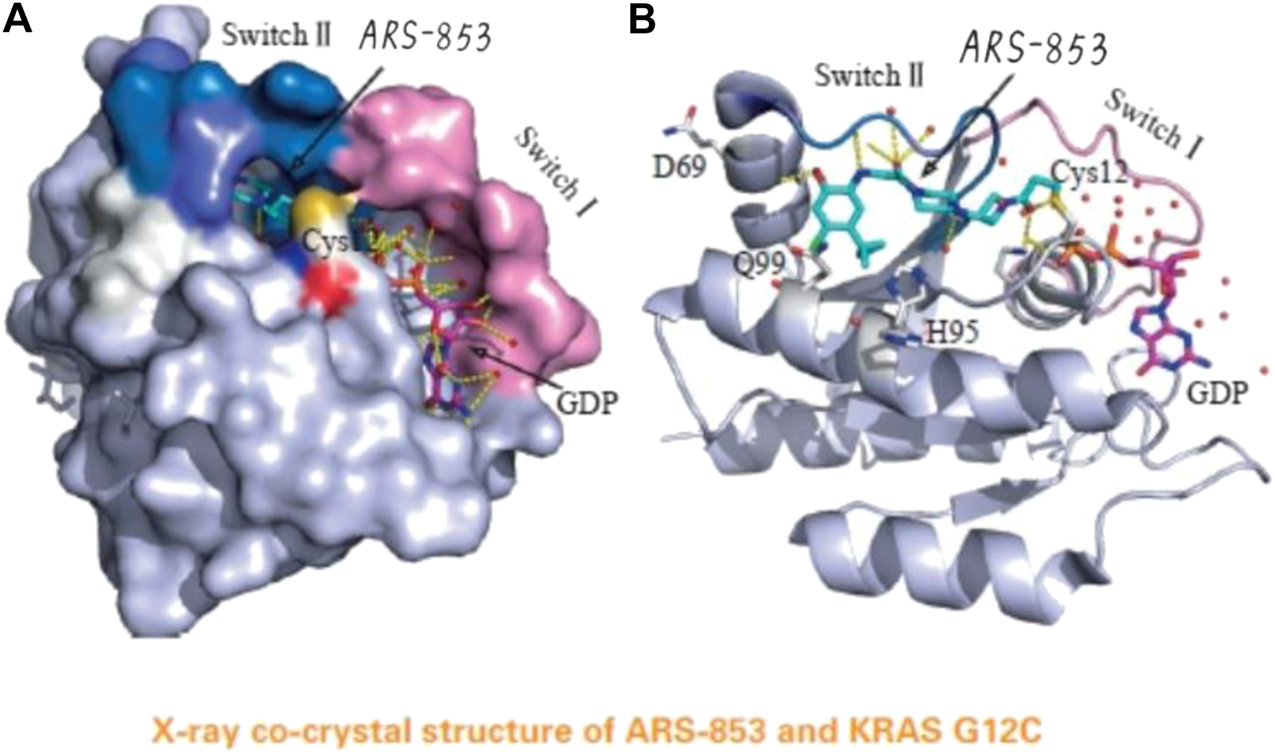
The acrylamide of ARS-853 can form covalent bond with 12 cysteine and extend to switch Ⅱ region, aromatic nucleus occupies hydrophobic region, the cyclopropyl group and the surrounding amino acids form a strong van der Waals force. (Reprinted by permission from Progrss in Pharmaceutical Sciences [14]).
FIGURE 5
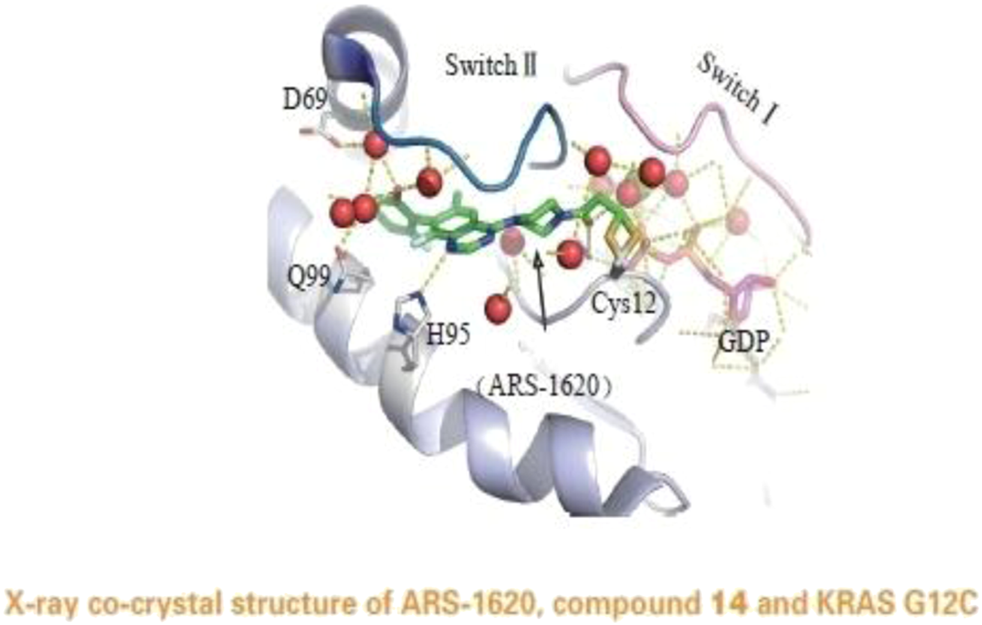
X-ray co-crystal structure of ARS-1620, compound 14 and KRAS G12C. (Reprinted by permission from Progrss in Pharmaceutical Sciences [14]).
FIGURE 6
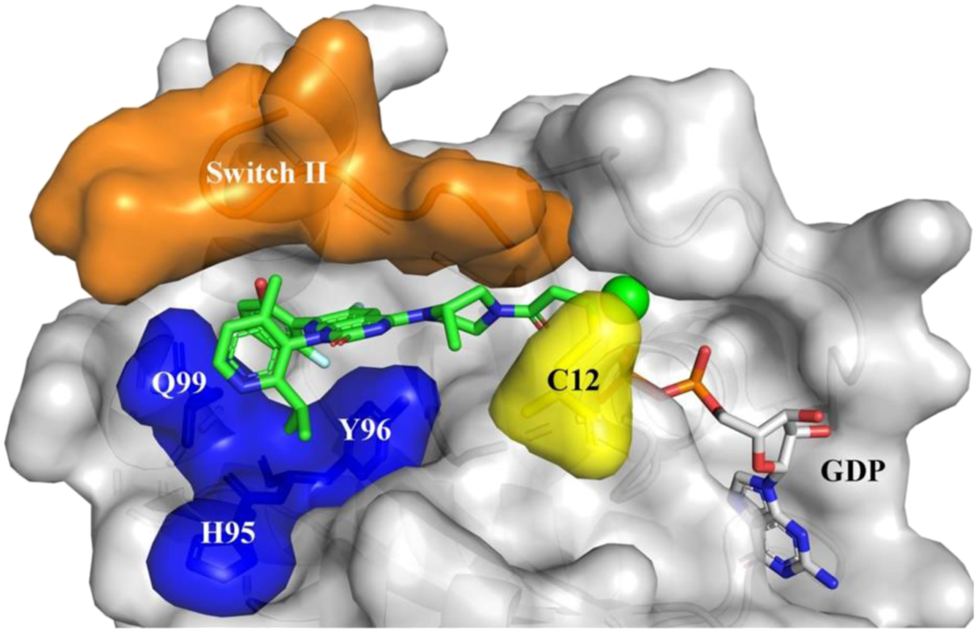
Sotorasib (AMG510) binding to KRAS-G12C protein. Orange: Switch II domain; Yellow: C12 residual; Blue: A cryptic pocket composed of H95, Y96, and Q99. AMG510 (green) and GDP (gray) are shown in sticks. (Reprinted by permission from Acta Pharmaceutica Sinica [20]).
FIGURE 7
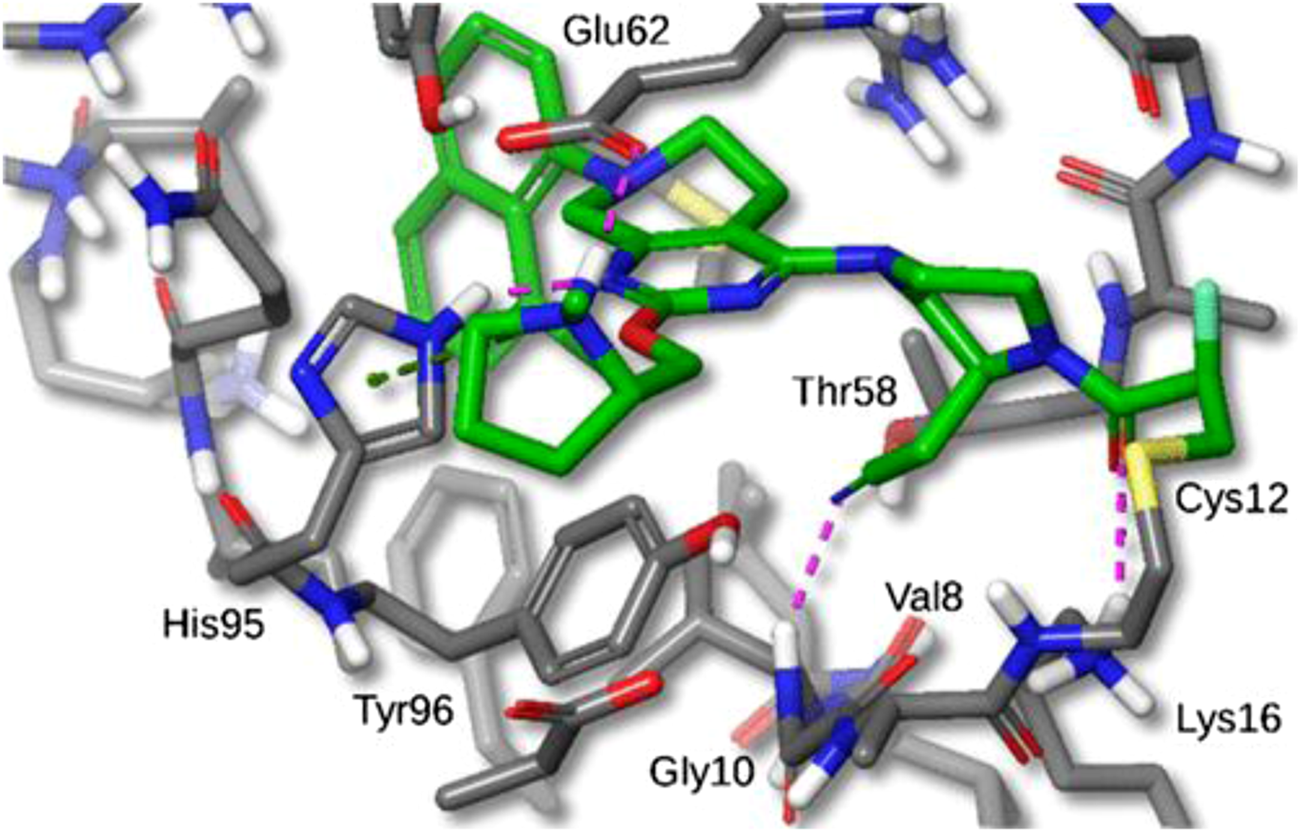
X-ray crystal structure of MRTX849 bound to KRASG12C with 1.94 Å resolution, hydrogens added for clarity. (Reprinted by permission from [30], further permissions related to the material excerpted should be directed to the ACS).
FIGURE 8
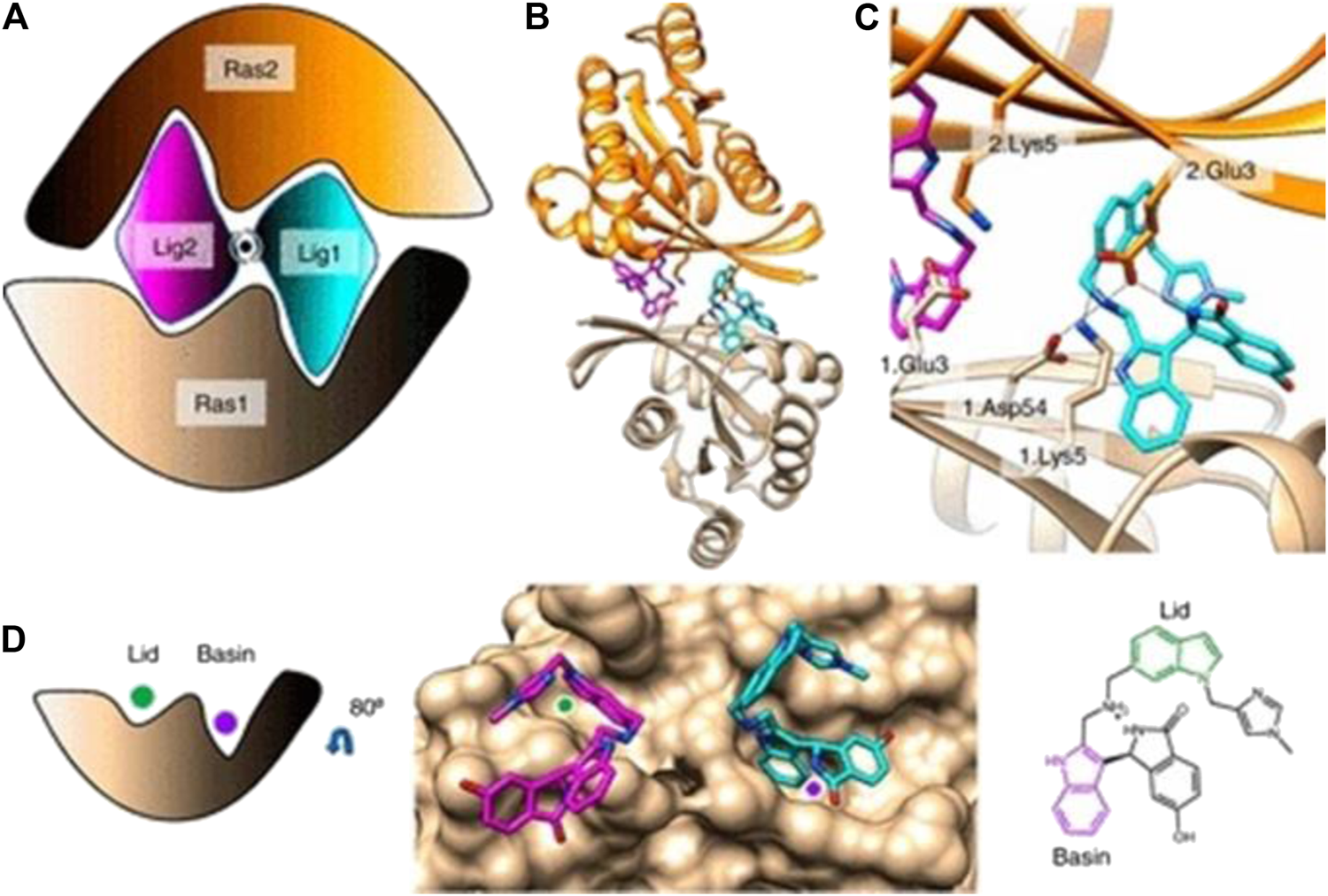
(A) Cartoon of BI-2852 KRAS dimer. Lig1 (cyan) and Lig2 (magenta) interact with both Ras1 (beige) and Ras2 (orange). (B) KRAS (ribbons) and key side chains are shown. (C) Zoom-in of the binding site: salt–bridge interactions shown (dashed lines). (D) Lid (green) and Basin (purple) are indicated on Ras1. Both Lig1 and Lig2 are shown on Ras1. The indole rings, colored green and purple, engage the Lid and Basin, respectively. (Reprinted by permission from Proc Natl Acad Sci U S A [33]).
The sources for the figures have been added to the reference list, and the reference numbering has been updated to accommodate this. The new references are appear below.
The authors apologize for this error and state that authorization has been obtained from the author of each picture, and this does not change the scientific conclusions of the article in any way. The original article has been updated.
References
3.
Prior IA Lewis PD and Mattos C. A Comprehensive Survey of Ras Mutations in Cancer. Cancer Res (2012) 72(10):2457–67. 10.1158/0008-5472.can-11-2612
4.
Simanshu DK Nissley DV and Mccormick F. RAS Proteins and Their Regulators in Human Disease. Cell (2017) 170(1):17.
14.
Huili L Fei J Jiwei R Tao L Yadong C. Research Advances on KRAS and Its Inhibitors. Acta Pharmaceutica Sinica (2020) 44(1):43–55.
20.
Xin L Yijun W and Pingyu L . Recent Advancement In Targeting The KRASG12C Mutant For Cancer Therapy. Acta Pharmaceutica Sinica (2021) 56(2):374–82.
30.
Fell JB Fischer JP Baer BR Blake JF Bouhana K Briere DM et al Identification of the Clinical Development Candidate MRTX849, a Covalent KRASG12C Inhibitor for the Treatment of Cancer. J Med Chem (2020) 63(13):6679–93. 10.1021/acs.jmedchem.9b02052
33.
Tran TH Alexander P Dharmaiah S Agamasu C and Balius TE . The Small Molecule BI-2852 Induces A Nonfunctional Dimer Of KRASProc National Academy Sci. (2020) 117(7).
Summary
Keywords
KRAS mutation, targeted drugs, oncogene, inhibitor, oncology
Citation
Yang A, Li M and Fang M (2022) Corrigendum: The Research Progress of Direct KRAS G12C Mutation Inhibitors. Pathol. Oncol. Res. 27:1610250. doi: 10.3389/pore.2021.1610250
Received
09 December 2021
Accepted
29 December 2021
Published
24 February 2022
Volume
27 - 2021
Edited and reviewed by
József Timar, Semmelweis University, Hungary
Updates
Copyright
© 2022 Yang, Li and Fang.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Min Li, doctorlimin@163.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.