Abstract
The neurotrophic tyrosine kinase receptor (NTRK) gene family is of rising importance as their fusions are oncogenic, and specific target drugs are available to inhibit the chimera proteins. Pan-TRK antibody, which shows the overexpression of the NTRK1-2-3 genes, is a useful tool to detect tumors with or without NTRK gene alterations, due to high negative predictive value. Though it is well known that pan-TRK immunopositivity is usually not connected to NTRK fusion, the role of other possible genetic alterations is under-researched. In our previous work, we found 3 NTRK1 amplified cases out of 6 cases with recurrent NTRK1 tyrosine kinase domain mutation pair, so we extended our investigation to a larger series to estimate amplification frequency. Pan-TRK immunopositivity was seen in 76 of the 132 dedifferentiated liposarcomas cases, followed by NTRK1-2-3 break-apart FISH tests in 76 pan-TRK positive cases to detect oncogenic fusions or other copy number alterations of these genes. None of the pan-TRK immunopositive dedifferentiated liposarcomas showed absolutely certain sign of fusion, however, 18 (28%) cases showed amplification of one of the genes, 13 had polysomy, 34 were normal, 11 were not evaluable. The extent of pan-TRK immunoreaction showed a positive correlation (p = 0.002) with the NTRK status found by FISH. Analyzing publicly available data from large series of 265 liposarcoma samples consisting of both well-differentiated and dedifferentiated liposarcoma case, 23 (8.6%) cases showed a mutual exclusive amplification of the NTRK genomic loci in a non-preselected, independent patient population indicating that our findings are presented in other cohorts. Our results underline the so far not revealed frequent occurrence of NTRK amplifications which might be important in the TRK inhibition therapy.
Introduction
Background
The role of NTRK fusion and overexpression of the TRK receptor has great clinical relevance in soft tissue sarcomas as targeted therapy option is available for those patients [1–4]. Extensive investigation has been performed to identify novel entities or tumor subtypes with the involvement of the NTRK genes.
NTRK gene and pan-TRK immunohistochemistry
The neurotrophic tyrosine kinase receptor (NTRK) gene family consists of three genes: NTRK1, NTRK2 and NTRK3 [5]. These genes are localized on different chromosomes, respectively: 1q23.1, 9q21.33, and 15q25.3 [5]. These genes encode three tyrosine kinase receptors (TRKA, TRKB and TRKC), that play a role in the regulation of apoptosis, survival, differentiation and proliferation in several cell types through well-known downstream signaling pathways, including PLCγ/PKC, MAPK/ERK and PI3K/AKT [5]. The tyrosine kinase receptors consist of extracellular, ligand-binding domain, transmembrane domain and intracellular, tyrosine kinase domain [5, 6]. The most important gene alterations affecting the NTRK gene family are fusions [6, 7]. NTRK fusions are characteristic in some rare tumors, like infantile fibrosarcoma [8], and congenital mesoblastic nephroma [9], but can occur in common malignancies as well, like lung cancer [10], colorectal cancer [11], melanoma [12], though less frequently. However, the fusions are the most investigated and best known, other gene alterations can occur in the NTRK genes. Recently, oncogenic point mutations within the extracellular, transmembrane and tyrosine kinase domains as well were identified in haematological malignancies [13, 14]. Furthermore, a deletion in the extracellular domain of TRKA showed oncogene capability in acute myeloid leukemia [15]. Also, a TRKA splice variant promoted tumorigenic cell behavior [16]. Although, NTRK1 amplification occurs in 8% of breast cancers [17], and several non-fusion NTRK alterations have been described in 14% of adult and pediatric tumors as well, their role in oncogenesis is yet to be explored [18, 19]. The NTRK gene family is at the center of attention nowadays, due to the highly effective, selective TRK inhibitors [20]. The US Food and Drug Administration approved the usage of specific TRK inhibitors, larotrectinib [1] and entrectinib [2], in case of any malignancy with NTRK fusion, regardless of the exact entity. Due to acquired resistance mutations within the tyrosine kinase domain, second-generation TRK inhibitors, repotrectinib [3] and selitrectinib [4], were developed.
Pan-TRK immunohistochemistry, which indicates the presence of TRKA and/or TRKB and/or TRKC protein(s), is a valuable tool to identify tumors without NTRK gene rearrangements, as it has a high negative predictive value [21]. Pan-TRK immunopositivity can be detected in some soft tissue sarcomas showing myogenic phenotype or myogenic differentiation, without NTRK fusion [22, 23].
In our previous work we investigated 131 dedifferentiated liposarcoma (DDLPS) cases and found 75 pan-TRK immunohistochemically positive cases [24]. Among the pan-TRK positive cases we found 6 cases with recurrent NTRK1 c.1810C>T (p.H604Y) and c.1838G>T (p.G613V) tyrosine kinase domain mutation pair and interestingly 3 of this 6 cases showed NTRK1 amplification. This surprisingly high frequency prompted us to extend our investigation and to estimate the real amplification frequency in this 75 cases. One additional pan-TRK positive case was included, so the total number of investigated cases were 76.
Our hypothesis was to investigate the relation between pan-TRK immunopositivity and possible genetic alterations by using NTRK1/2/3 locus specific break apart probe set in order to estimate the frequency of genomic rearrangement and/or regional or overall copy number alterations related to any of these three regions.
Materials and methods
Case selection
We analyzed 76 individual cases of dedifferentiated liposarcoma with myogenic heterologous differentiation and pan-TRK immunopositivity from our institutional and consultation archives (diagnosed between 2014 and 2024) in this study. The diagnosis was based on clinical features, characteristic morphology and immunohistochemical and/or molecular examination of MDM2 and/or CDK4 overexpression or amplification. Previously executed immunohistochemistry with desmin and alpha smooth muscle actin antibody were used to show myogenic heterologous differentiation, and pan-TRK antibody was used to detect NTRK1-2-3 overexpression. This study was approved by the Semmelweis University Regional and Institutional Committee of Science and Research Ethics (TUKEB 155/2012).
Immunohistochemistry (IHC)
Immunohistochemistry was performed on deparaffinized, rehydrated whole slide formalin-fixed, paraffin-embedded (FFPE) block sections with the Bond-Max automated staining system (Leica Biosystems, Wetzlar, Germany), after applying antibody-specific epitope retrieval techniques, using the following antibody: pan-TRK (ready-to-use, EPR17341, Ventana). The intensity and extent of immunoreactivity were evaluated by two soft tissue pathologists (ZS and KD). Tumor samples were considered positive with pan-TRK antibody if >1% of tumor cells exhibited immunostaining at any intensity above the background. Staining intensities of tumor cells were graded as no staining (0), very weak (1), weak (2), moderate (3), strong (4) or very strong (5) and numbers of stained tumor cells as the extent of staining were graded as 0% (0), 1%–19% (1), 20%–39% (2) 40%–59% (3), 60%–79% (4), 80%–100% (5).
Fluorescence in situ hybridization (FISH)
FISH was performed on interphase nuclei from 3 µm sections of FFPE blocks using the ZytoLight SPEC Dual Color Break Apart Probes of the NTRK1/2/3 genes (ZytoVision, Bremerhaven, Germany). The slides were analyzed by assessing a minimum of 100 cells each in a tumor region of interest. Rearrangement of the NTRK genes was defined as a minimum of 20% tumor cells with split signals (classical pattern) of the corresponding NTRK probes. Polysomy or copy number gain was defined by an average copy number of all NTRK (1/2/3) signals ≥3. Amplification was defined if a NTRK gene showed at least twice in number of signals compared to the average of the other two NTRK gene type. In other words, the different NTRKs (locating on different chromosomes) served as internal controls to assess whether there is a polysomy or either of them is amplified. Because we used break-apart NTRK probes, we could distinguish “whole” amplification (the whole gene was affected) or “partial” amplification (either the 3′ end/red signal or the 5′ end/green signal was amplified). FISH signals were scored by one pathologist (ZS) and one biologist (GP).
Next-generation sequencing (NGS)
Targeted DNA and RNA libraries were prepared according to the TruSight Tumor 170 reference guide (Illumina, San Diego, CA, United States). The TruSight Tumor 170 kit included the analysis of 151 genes for insertion, deletion, single nucleotide variant, copy number variation and 55 genes for fusions and splice variants and 59 genes for amplification. Genomic DNA and total RNA were isolated from formalin-fixed, paraffin-embedded specimens using the QIAamp® DNA FFPE Tissue Kit (Qiagen) and the High Pure FFPET RNA Isolation Kit (Roche Diagnostics, Mannheim, Germany). Estimations of tumor cell percentages of the samples were performed by histopathological examinations prior to the isolation processes. DNA and RNA concentrations were measured using the Qubit™ dsDNA HS Assay and Qubit™ RNA HS Assay Kits (Thermo Fisher Scientific) on a Qubit™ 4 Fluorometer. 120 ng DNA in 52 μL volume was sheared (200 cycles, peak power: 75 W, duty factor: 10, treatment time: 510 s, at 7°C setpoint) using a Covaris M220 Focused-ultrasonicator (Covaris, Woburn, United States). 80 ng of DNase digested RNA was used for library preparation of each sample The size of double-stranded DNA fragments and RNA molecules was confirmed after shearing using a Tapestation 4,200 (Agilent, Cheshire, UK). Library preparation workflow of Illumina TruSight Tumor 170 assay was performed according to the manufacturer’s protocol. The final libraries were paired-end sequenced at a 2 × 101 bp read length, using Illumina NextSeq 1000/2000 P3 (200 cycles) reagent kits on a NextSeq 2000 platform. Bioinformatic analysis was performed using TruSight Tumor 170 v2.0.2 Local App (Illumina, San Diego, CA, United States). For further clinical interpretation, we used Genoox’s Franklin web-based analysis tool, which applies variant filtering and further annotations: pathogenicity scores, population-frequency, protein structure predictions, relevant clinical guidelines and therapeutic options for variants.
In silico analysis
Publicly available data sets from TCGA and cBioPortal were analysed for the presence of structural variants involving NTRK1/2/3 gene loci. From three studies analyzing soft tissue sarcomas, well-differentiated and dedifferentiated liposarcoma cases were further analyzed to interrogate the frequencies of NTRK1/2/3 genes and MDM2 and CDK4 genes. From the 2551 cases presented in these three studies 265 unique well-differentiated or dedifferentiated liposarcoma samples were included for the analysis using cBioPortal [25–29].
Statistical analysis
NTRK gene family statuses by FISH (normal, polyploidy and amplification) were investigated based on pan-TRK immunohistochemistry (extent and intensity values). In order to compare three samples, one can use ANOVA test; however, in this case, there are two assumptions to be justified: 1. Normality of residuals (the errors used for the estimation of the error terms are normally distributed), 2. Homogeneity of variance (the level of variance for a particular variable is constant across the sample). For testing normality, we used one-sample Kolmogorov-Smirnov normal test; and Levene Statistic was used to test homogeneity of variances. Finally, to compare normal, polysomic and amplification groups, parametric one-way ANOVA and non-parametric Kruskal-Wallis tests were run.
Results
We included 76 pan-TRK immunpositive dedifferentiated liposarcoma cases in our investigation. The diagnosis of dedifferentiated liposarcoma was based on histology and MDM2 and/or CDK4 immunohistochemistry findings (Figure 1). In cases with unequivocal immunohistochemistry results, FISH examinations were executed with MDM2 and/or CDK4 probes. The male-female ratio is 1.24 (42:34). The most common site for dedifferentiated liposarcoma in our setting was retroperitoneum (42%). Basic clinical data is available in an article of our previous study [24].
FIGURE 1
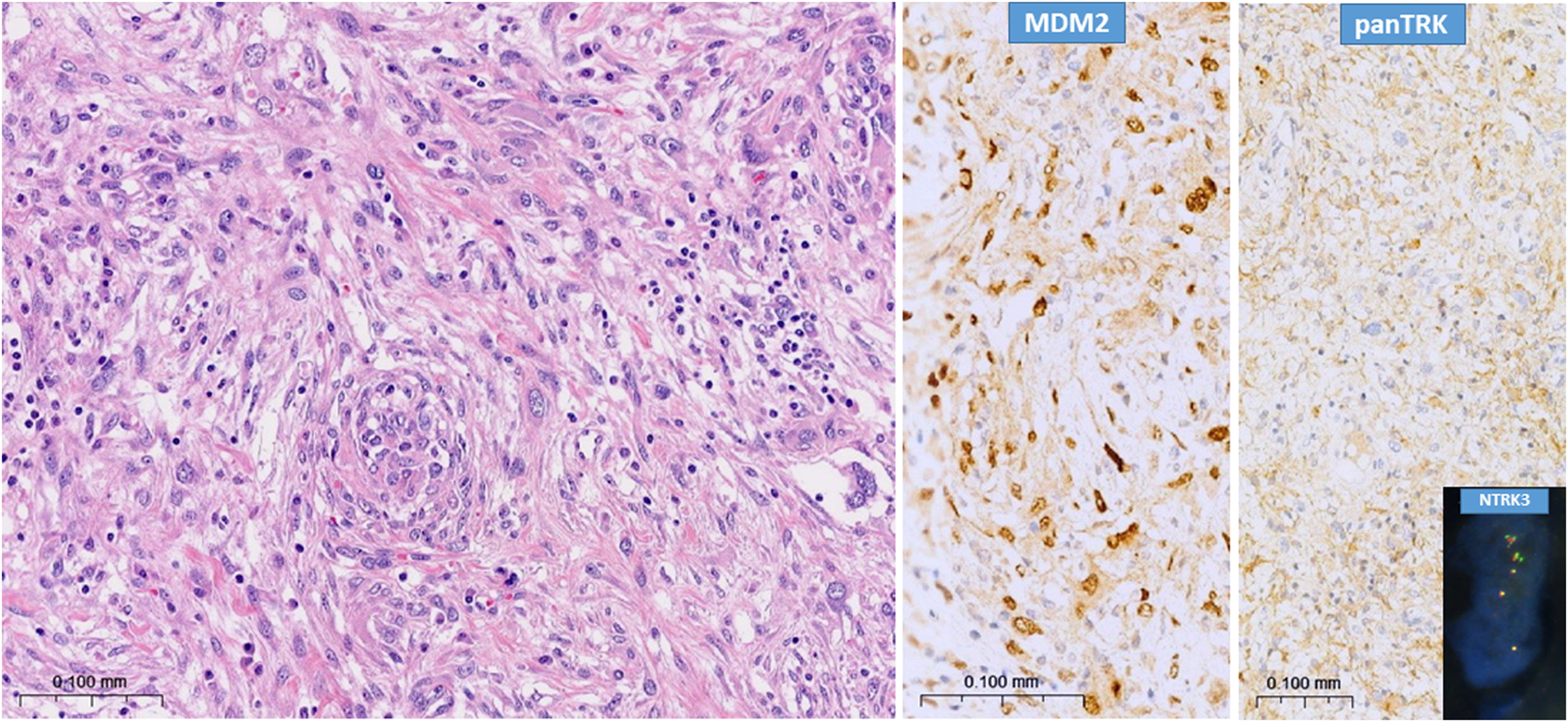
Typical picture of dedifferentiated liposarcoma (H&E) with characteristic strong MDM2 nuclear immunopositivity and strong pan-TRK cytoplasmic positivity. Insert shows NTRK3 amplification (break-apart probe), case N°113.
With NTRK1-2-3 FISH probes, among the 76 pan-TRK immunopositive cases, 34 proved to be normal as no copy number gain was evinced in the NTRK gene family. 13 cases proved to have polysomy because of the involvement of all three NTRK genes, which are localized on different chromosomes, showed increased number of signals. Mutually exclusive amplification pattern was seen in 18 cases (NTRK1: 9; NTRK2: 5; NTRK3: 4). 5 of these showed partial amplification since only the 3′ or the 5′ region of the given gene was amplified, while the other 13 cases showed the amplification of the whole sampled locus, as both, co-localised signals of the tested gene locus showed an increased copy number (Figure 2; Table 1). No classical NTRK fusion was detected by FISH. The partial amplification pattern is suggestive of fusion with unknown partners. To test possible fusions, we performed NGS (TruSight Tumor 170 kit) in one case of partial amplification, but no fusion was detected. Unfortunately, 11 cases were not evaluable in regard to NTRK gene family status due to FISH technical issues or insufficient amount of tumor material (Figures 1, 2; Table 1; Supplementary Table S1).
FIGURE 2
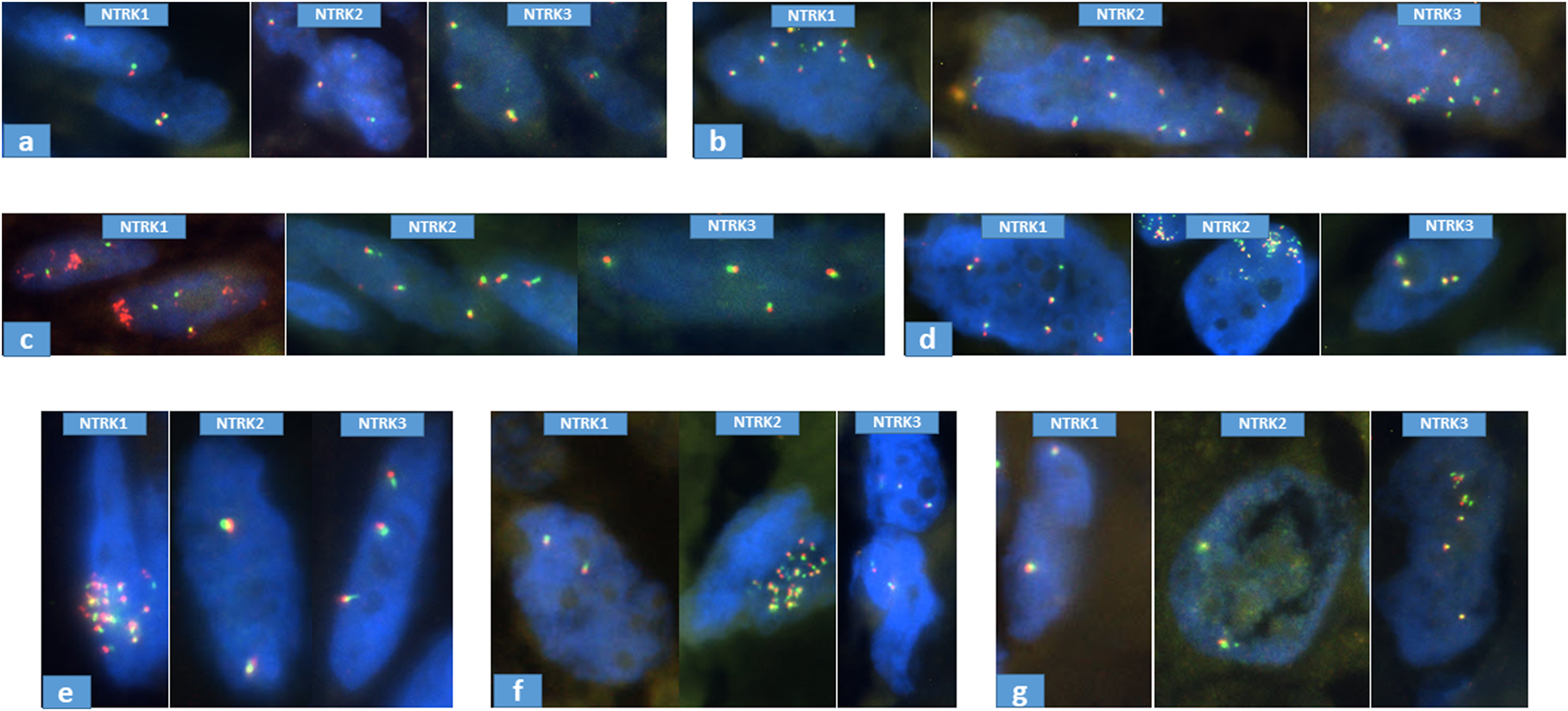
Representative samples of NTRK FISH results. (A): normal, case N°20; (B): polysomy, case N°85; (C): partial amplification of NTRK1 (3′), case N°7; (D): partial amplification of NTRK2 (5′), case N°115; (E): whole amplification of NTRK1, case N°84; (F): whole amplification of NTRK2, case N°11; (G): whole amplification of NTRK3, case N°113.
TABLE 1
| Patient ID | NTRK gene | FISH result | Copy number | Comment | Interpretation | Affected gene |
|---|---|---|---|---|---|---|
| 7 | NTRK1 | extra red | green sign 3.8; red sign >10 | NTRK1 3′ region amplification | amplification | NTRK1 |
| NTRK2 | negative | 4 | ||||
| NTRK3 | negative | 3 | ||||
| 8 | NTRK1 | negative | 3.1 | amplification | NTRK2 | |
| NTRK2 | negative | 6.5 | occasionally >10 sign | |||
| NTRK3 | negative | 3.2 | ||||
| 11 | NTRK1 | negative | 2.2 | amplification | NTRK2 | |
| NTRK2 | negative | 7.4 | occasionally >10 sign | |||
| NTRK3 | negative | 2.2 | ||||
| 23 | NTRK1 | negative | 5.5 | occasionally >10 sign | amplification | NTRK1 |
| NTRK2 | negative | 2.1 | ||||
| NTRK3 | negative | 2.4 | ||||
| 44 | NTRK1 | negative | 2 | amplification | NTRK3 | |
| NTRK2 | negative | 2 | ||||
| NTRK3 | negative | 5.2 | ||||
| 123 | NTRK1 | negative | 3.1 | amplification | NTRK3 | |
| NTRK2 | negative | 3.1 | ||||
| NTRK3 | extra green | green sign 6.2; red sign 2.9 | NTRK3 5′ region amplification | |||
| 129 | NTRK1 | negative | 2.2 | amplification | NTRK2 | |
| NTRK2 | negative | 5.8 | ||||
| NTRK3 | negative | 3.1 | ||||
| 115 | NTRK1 | negative | 8.2 | amplification | NTRK2 | |
| NTRK2 | extra green | green sign >10; red sign ∼8 | NTRK2 5′ region amplification | |||
| NTRK3 | negative | 7.5 | ||||
| 112 | NTRK1 | extra red | green sign 2.4; red sign >10 | NTRK1 3′ region amplification | amplification | NTRK1 |
| NTRK2 | negative | 3.2 | ||||
| NTRK3 | negative | 4.8 | ||||
| 113 | NTRK1 | negative | 3.5 | amplification | NTRK3 | |
| NTRK2 | negative | 3.6 | ||||
| NTRK3 | negative | >10 | whole NTRK3 region amplification | |||
| 121 | NTRK1 | negative | 2.2 | amplification | NTRK2 | |
| NTRK2 | extra red | green sign 2; red sign >10 | NTRK2 3′ region amplification | |||
| NTRK3 | negative | 2.7 | ||||
| 82 | NTRK1 | negative | 2.5 | amplification | NTRK3 | |
| NTRK2 | negative | 2 | ||||
| NTRK3 | negative | >10 | whole NTRK3 region amplification | |||
| 86 | NTRK1 | negative | 9.5 | whole NTRK1 region amplification | amplification | NTRK1 |
| NTRK2 | negative | 3.2 | ||||
| NTRK3 | negative | 3.1 | ||||
| 51 | NTRK1 | negative | >10 | whole NTRK1 region amplification | amplification | NTRK1 |
| NTRK2 | negative | 3.4 | ||||
| NTRK3 | negative | 3.2 | ||||
| 132 | NTRK1 | negative | >10 | whole NTRK1 region amplification | amplification | NTRK1 |
| NTRK2 | negative | 2 | no fusion by NGS | |||
| NTRK3 | negative | 2.2 | ||||
| 84 | NTRK1 | negative | >10 | whole NTRK1 region amplification | amplification | NTRK1 |
| NTRK2 | negative | 3.8 | no fusion by NGS | |||
| NTRK3 | negative | 3.5 | ||||
| 88 | NTRK1 | negative | 8.5 | whole NTRK1 region amplification | amplification | NTRK1 |
| NTRK2 | negative | 2 | no fusion by NGS | |||
| NTRK3 | negative | 2 | ||||
| 125 | NTRK1 | negative | >10 | whole NTRK1 region amplification | amplification | NTRK1 |
| NTRK2 | negative | 2 | no fusion by NGS | |||
| NTRK3 | negative | 2.5 |
NTRK1-2-3 break-apart FISH results of amplified cases.
The intensity and the extent of pan-TRK immunoreactions varied over a wide range (Table 2; Supplementary Table S2; Supplementary Table S3).
TABLE 2
| NTRK gene family status by FISH | Pan-TRK immunohistochemistry | ||
|---|---|---|---|
| Score | Extent | Intensity | |
| normal (n = 34) | 1 | 13 | 28 |
| 2 | 6 | 1 | |
| 3 | 5 | 4 | |
| 4 | 3 | 1 | |
| 5 | 7 | 0 | |
| average | 2.56 | 1.35 | |
| polysomy (n = 13) | 1 | 2 | 8 |
| 2 | 1 | 2 | |
| 3 | 2 | 2 | |
| 4 | 2 | 0 | |
| 5 | 6 | 1 | |
| average | 3.69 | 1.77 | |
| amplification (n = 18) | 1 | 3 | 10 |
| 2 | 0 | 3 | |
| 3 | 1 | 3 | |
| 4 | 1 | 0 | |
| 5 | 13 | 2 | |
| average | 4.17 | 1.94 | |
Detailed pan-TRK immunohistochemistry results of dedifferentiated liposarcoma cases with different NTRK gene family status.
265 samples consisting of 38 well-differentiated and 227 dedifferentiated liposarcoma cases were retrieved from cBioPortal. Amplification of the NTRK1/2/3 genes were seen in 23/265 (8.6%) of the reported cases without preselection using pan-TRK immunohistochemistry for NTRK1/2/3. From these, 9/265 (3.3%), 11/265 (4.1%) and 3/265 (1.1%) cases showed amplification of the NTRK1, NTRK2 and NTRK3 genes, respectively. In all cases a mutually exclusive amplification pattern was observed. Only two cases (1 NTRK1 and 1 NTRK3) with amplification were seen in well-differentiated liposarcoma samples. Single nucleotide variants (SNVs) of the NTRK1/2/3 genes were reported in three cases, NTRK1 G619R, NTRK2 S180G and NTRK3 D167H. Intriguingly, the NTRK1 G619R was in a case with NTRK2 amplification, and the NTRK3 D167H was in a case with NTRK1 amplification.
Statistics for FISH and immunohistochemistry results
The statistical analysis showed that in all groups (normal, polysomy and amplification) the assumption of normality of the residues is not met, neither in terms of extent nor in terms of intensity. The homogeneity of variance was found to be fulfilled in terms of the extent. Since simulation studies using a variety of non-normal distributions have shown that the false positive rate is not affected very much by the violation of the normality assumption [30, 31], we used one-way ANOVA to investigate extent. It resulted in p = 0.002, which means that the groups are significantly different, moreover post hoc test showed the following p values: p = 0.075 for normal and polysomy groups, p = 0.684 for polysomy and amplification groups, and p = 0.02 for normal and amplification groups. The conclusion is that normal and amplification groups are significantly different in term of extent. In term of intensity, the homogeneity of variance was found to not be fulfilled, thus instead of ANOVA, non-parametric Kruskal-Wallis test was used. It resulted in p = 0.116 meaning that we should retain the null hypothesis, namely that there is no significant difference between the three groups in term of intensity, however, the tendency for increased intensity from normal to amplification group can be established (Table 2).
Discussion
As pan-TRK immunohistochemistry has a high negative predictive value, only pan-TRK immunopositive cases were investigated with NTRK probes to find NTRK gene alterations.
We studied 132 dedifferentiated liposarcoma cases after pan-TRK immunostaining by using NTRK1/2/3 locus specific break-apart probe set. FISH was successfully performed in 65 of the 76 pan-TRK IHC preselected cases, and amplification patterns of one of the NTRK genes was seen in 18 cases (27.6%). A partial amplification pattern, meaning that only one of the two break apart FISH probes showed amplification was observed in 5 cases. The partial amplification pattern might be compatible with fusion of the involved genes, similar to cases described in dermatofibrosarcoma protuberans, with COL1A1::PDGFB or in case of EWSR1::NFATC2 fusion (EWSR1 pattern of FISH) in round cell sarcomas with EWSR1–non-ETS fusions [32]. Therefore, we performed NGS of one of these cases (case number: 112), but there was no NTRK fusion identified. Full transcriptome sequencing or whole genome sequencing might be needed to detect rare NTRK fusions with this type of FISH pattern. So far, there is only a limited literature on dedifferentiated liposarcomas harboring NTRK fusions. In a letter format, the authors reported two cases of dedifferentiated liposarcomas with NTRK fusions, though they admit that the functional significance was not clinically demonstrated” [33]. In a study trying to find NTRK fusion in solid tumors, one retroperitoneal liposarcoma showed double NTRK3 fusions (MORF4L1:NTRK3 and PPFIA2:NTRK3). Only 40 cases showed NTRK fusions among the investigated 10.194 patients with solid tumors (0.4%), and it was most frequently identified in soft tissue sarcomas (3.0%). Among soft tissue sarcomas, the fibrosarcoma subtype had an exceptionally high prevalence for NTRK fusions (12.7%) [34].
We found 18 cases with mutually exclusive amplification NTRK genomic loci. To our knowledge, the link between NTRK amplification and dedifferentiated liposarcoma was not investigated directly ever before. As 18/65 (27.6%) of the selected cases showed an amplification of in one of the NTRK genes, we further searched publicly available data derived from liposarcoma samples (both dedifferentiated and well-differentiated liposarcoma cases) from cBioPortal to estimate the frequency in an unbiased sample set. Amplification of the NTRK genes were observed 23/265 (8.6%) samples. This observed frequency is much lower than those observed in our study, however, here samples were analyzed without preselection for the presence of NTRK expression by IHC. Soft tissue sarcomas were screened for NTRK gene alterations other than fusions only in a few research [33–36]. NTRK amplifications were found in some tumors, like biliary tract cancers [37] and non-small cell lung cancer [38].
Unsurprisingly, as the focus of researches is still on fusions, the correlation of pan-TRK immunohistochemistry and NTRK amplification was not clarified before. Among 27 NTRK amplified tumors with various histology types only 4 showed positive pan-TRK immunostaining [39]. Two cases of soft tissue spindle cell tumors with NTRK fusion and co-occurring amplification as well showed positive pan-TRK immunoreaction [35]. Pan-TRK immunopositivity was seen in two cases of gastric cancers harboring NTRK amplifications [40].
There is no literature of partial NTRK amplification in tumors, and neither of polysomy in dedifferentiated liposarcoma at all, to our knowledge. However, polysomy and/or amplification of some genes (i.e., ALK and HER2) may indicate therapy against the tumors harboring these genes. In this way further investigation is needed whether it can be applicate in cases of DDLPSs with NTRK polysomy or amplification.
Viewing the NTRK gene family status by FISH, comparing it with pan-TRK immunohistochemistry, we can summarize that tumors with NTRK amplification are more likely to have a stronger and more diffuse pan-TRK immunoreaction, than those without NTRK rearrangement. Furthermore, cases with polysomy as well have a higher tendency to express a wider and more intense pan-TRK immunoreaction, but falling behind the NTRK amplified ones in that regard.
The relation between the gain of these genomic loci and increased expression of the gene regarding functional activity of the involved receptors requires further investigation.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The studies involving humans were approved by the Semmelweis University Regional and Institutional Committee of Science and Research Ethics (TUKEB 155/2012). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin because we dealt only with paraffin-embedded material.
Author contributions
ZL, GP, and ZS conducted the experiments, ZL, KS, JS, and ZS wrote the manuscript, and DK contributed in the pathological evaluation. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.por-journal.com/articles/10.3389/pore.2024.1611993/full#supplementary-material
References
1.
Doebele RC Davis LE Vaishnavi A Le AT Estrada-Bernal A Keysar S et al An oncogenic NTRK fusion in a patient with soft-tissue sarcoma with response to the tropomyosin-related kinase inhibitor LOXO-101. Cancer Discov (2015) 5(10):1049–57. 10.1158/2159-8290.CD-15-0443
2.
Farago AF Le LP Zheng Z Muzikansky A Drilon A Patel M et al Durable clinical response to entrectinib in NTRK1-rearranged non-small cell lung cancer. J Thorac Oncol (2015) 10(12):1670–4. 10.1097/01.JTO.0000473485.38553.f0
3.
Drilon A Ou SHI Cho BC Kim DW Lee J Lin JJ et al Repotrectinib (TPX-0005) is a next-generation ROS1/TRK/ALK inhibitor that potently inhibits ROS1/TRK/ALK solvent- front mutations. Cancer Discov (2018) 8(10):1227–36. 10.1158/2159-8290.CD-18-0484
4.
Drilon A Nagasubramanian R Blake JF Ku N Tuch BB Ebata K et al A next-generation TRK kinase inhibitor overcomes acquired resistance to prior TRK kinase inhibition in patients with TRK fusion-positive solid tumors. Cancer Discov (2017) 7(9):963–72. 10.1158/2159-8290.CD-17-0507
5.
Lippai Z Sapi Z . Diagnostic approach of tumors with neurotrophic tropomyosin receptor tyrosine kinase gene fusions. Orv Hetil (2020) 161(41):1753–63. 10.1556/650.2020.31846
6.
Vaishnavi A Le AT Doebele RC . TRKing down an old oncogene in a new era of targeted therapy. Cancer Discov (2015) 5(1):25–34. 10.1158/2159-8290.CD-14-0765
7.
Solomon JP Benayed R Hechtman JF Ladanyi M . Identifying patients with NTRK fusion cancer. Ann Oncol (2019) 30(Suppl. 8):viii16–viii22. 10.1093/annonc/mdz384
8.
Davis JL Lockwood CM Albert CM Tsuchiya K Hawkins DS Rudzinski ER . Infantile NTRK-associated mesenchymal tumors. Pediatr Dev Pathol (2018) 21(1):68–78. 10.1177/1093526617712639
9.
Zhao M Yin M Kuick CH Chen H Aw SJ Merchant K et al Congenital mesoblastic nephroma is characterised by kinase mutations including EGFR internal tandem duplications, the ETV6-NTRK3 fusion, and the rare KLHL7-BRAF fusion. Histopathology (2020) 77(4):611–21. 10.1111/his.14194
10.
Harada G Santini FC Wilhelm C Drilon A . NTRK fusions in lung cancer: from biology to therapy. Lung Cancer (2021) 161:108–13. 10.1016/j.lungcan.2021.09.005
11.
Ratti M Grizzi G Passalacqua R Lampis A Cereatti F Grassia R et al NTRK fusions in colorectal cancer: clinical meaning and future perspective. Expert Opin Ther Targets (2021) 25(8):677–83. 10.1080/14728222.2021.1978070
12.
Forschner A Forchhammer S Bonzheim I . NTRK gene fusions in melanoma: detection, prevalence and potential therapeutic implications. J Dtsch Dermatol Ges (2020) 18(12):1387–92. 10.1111/ddg.14160
13.
Joshi SK Qian K Bisson WH Watanabe-Smith K Huang A Bottomly D et al Discovery and characterization of targetable NTRK point mutations in hematologic neoplasms. Blood (2020) 135(24):2159–70. 10.1182/blood.2019003691
14.
Iacobucci I Wen J Meggendorfer M Choi JK Shi L Pounds SB et al Genomic subtyping and therapeutic targeting of acute erythroleukemia. Nat Genet (2019) 51(4):694–704. 10.1038/s41588-019-0375-1
15.
Reuther GW Lambert QT Caligiuri MA Der CJ . Identification and characterization of an activating TrkA deletion mutation in acute myeloid leukemia. Mol Cell Biol (2000) 20(23):8655–66. 10.1128/mcb.20.23.8655-8666.2000
16.
Tacconelli A Farina AR Cappabianca L Desantis G Tessitore A Vetuschi A et al TrkA alternative splicing: a regulated tumor-promoting switch in human neuroblastoma. Cancer Cell (2004) 6(4):347–60. 10.1016/j.ccr.2004.09.011
17.
Stravodimou A Voutsadakis IA . Neurotrophic receptor tyrosine kinase family members in secretory and non-secretory breast carcinomas. World J Clin Oncol (2022) 13(2):135–46. 10.5306/wjco.v13.i2.135
18.
Okamura R Boichard A Kato S Sicklick JK Bazhenova L Kurzrock R . Analysis of NTRK alterations in pan-cancer adult and pediatric malignancies: implications for NTRK-targeted therapeutics. JCO Precis Oncol (2018) 2018:1–20. 10.1200/PO.18.00183
19.
Hsiao SJ Zehir A Sireci AN Aisner DL . Detection of tumor NTRK gene fusions to identify patients who may benefit from tyrosine kinase (TRK) inhibitor therapy. J Mol Diagn (2019) 21(4):553–71. 10.1016/j.jmoldx.2019.03.008
20.
Kiss E Papai Z . Novel targeted therapeutic option in oncology: tropomyosin receptor tyrosine kinase inhibitors. Orv Hetil (2021) 162(34):1362–9. 10.1556/650.2021.32183
21.
Rudzinski ER Lockwood CM Stohr BA Vargas SO Sheridan R Black JO et al Pan-trk immunohistochemistry identifies NTRK rearrangements in pediatric mesenchymal tumors. Am J Surg Pathol (2018) 42(7):927–35. 10.1097/PAS.0000000000001062
22.
Chiang S Cotzia P Hyman DM Drilon A Tap WD Zhang L et al NTRK fusions define a novel uterine sarcoma subtype with features of fibrosarcoma. Am J Surg Pathol (2018) 42(6):791–8. 10.1097/PAS.0000000000001055
23.
Hung YP Fletcher CDM Hornick JL . Evaluation of pan-TRK immunohistochemistry in infantile fibrosarcoma, lipofibromatosis-like neural tumour and histological mimics. Histopathology (2018) 73(4):634–44. 10.1111/his.13666
24.
Lippai Z Péterfia B Papp G Dezső K Bedics G Pápai Z et al A recurrent NTRK1 tyrosine kinase domain mutation pair is characteristic in a subset of dedifferentiated liposarcomas. Eur J Cancer (2024) 202:114005. 10.1016/j.ejca.2024.114005
25.
Cerami E Gao J Dogrusoz U Gross BE Sumer SO Aksoy BA et al The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data (vol 2, pg 401, 2012). Cancer Discov (2012) 2(10):960.
26.
Gao J Aksoy BA Dogrusoz U Dresdner G Gross B Sumer SO et al Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal (2013) 6(269):pl1. 10.1126/scisignal.2004088
27.
Lazar AJ , Cancer Genome Atlas Research Network. Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell (2017) 171(4):950–65.e28. 10.1016/j.cell.2017.10.014
28.
Nacev BA Sanchez-Vega F Smith SA Antonescu CR Rosenbaum E Shi H et al Clinical sequencing of soft tissue and bone sarcomas delineates diverse genomic landscapes and potential therapeutic targets. Nat Commun (2022) 13(1):3405. 10.1038/s41467-022-30453-x
29.
Barretina J Taylor BS Banerji S Ramos AH Lagos-Quintana M Decarolis PL et al Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat Genet (2010) 42(8):715–21. 10.1038/ng.619
30.
Harwell MR . Summarizing monte-carlo results in methodological research. J Educ Stat (1992) 17(4):297–313. 10.2307/1165126
31.
Lix LM Keselman JC Keselman HJ . Consequences of assumption violations revisited: a quantitative review of alternatives to the one-way analysis of variance F test. Rev Educ Res (1996) 66(4):579–619. 10.2307/1170654
32.
Szuhai K Ijszenga M de Jong D Karseladze A Tanke HJ Hogendoorn PCW . The NFATc2 gene is involved in a novel cloned translocation in a Ewing sarcoma variant that couples its function in immunology to oncology. Clin Cancer Res (2009) 15(7):2259–68. 10.1158/1078-0432.CCR-08-2184
33.
Brahmi M Dufresne A Verret B Tirode F Blay JY . NTRK fusion in soft tissue sarcomas harboring MDM2/CDK4 amplification: three case reports. Ann Oncol (2021) 32(6):813–4. 10.1016/j.annonc.2021.02.019
34.
Xu Y Shi X Wang W Zhang L Cheung S Rudolph M et al Prevalence and clinico-genomic characteristics of patients with TRK fusion cancer in China. NPJ Precis Oncol (2023) 7(1):75. 10.1038/s41698-023-00427-3
35.
Suurmeijer AJ Dickson BC Swanson D Zhang L Sung YS Huang HY et al The histologic spectrum of soft tissue spindle cell tumors with NTRK3 gene rearrangements. Genes Chromosomes Cancer (2019) 58(11):739–46. 10.1002/gcc.22767
36.
Vargas AC Joy C Maclean FM Bonar F Wong DD Gill AJ et al Kinase expression in angiomatoid fibrous histiocytoma: panTRK is commonly expressed in the absence of NTRK rearrangement. J Clin Pathol (2024) 77(4):251–4. 10.1136/jcp-2023-209225
37.
Rizzato M Brignola S Munari G Gatti M Dadduzio V Borga C et al Prognostic impact of FGFR2/3 alterations in patients with biliary tract cancers receiving systemic chemotherapy: the BITCOIN study. Eur J Cancer (2022) 166:165–75. 10.1016/j.ejca.2022.02.013
38.
Li D Liu J Zhang X Han J Jin H Wang L et al Combined lorlatinib, dabrafenib, and trametinib treatment for ROS1-rearranged advanced non-small-cell lung cancer with a lorlatinib-induced braf V600E mutation: a case report. Cancer Manag Res (2022) 14:3175–9. 10.2147/CMAR.S387211
39.
Lee SJ Kim NK Lee SH Kim ST Park SH Park JO et al Gene amplification in patients with metastatic cancer. Precision Future Med (2017) 1(3):129–37.
40.
Xu AD Fu Y Pu XH Wu HY Sun Q Fan XS . Clinicopathological features of gastric carcinomas with NTRK-rearrangement/amplification: report of four cases. Zhonghua Bing Li Xue Za Zhi (2023) 52(5):454–9. 10.3760/cma.j.cn112151-20221008-00840
Summary
Keywords
soft tissue sarcoma, dedifferentiated liposarcoma, gene rearrangement, gene amplification, NTRK
Citation
Lippai Z, Papp G, Szuhai K, Sápi J, Dezső K and Sápi Z (2025) NTRK amplification occurs frequently in pan-TRK immunopositive dedifferentiated liposarcomas. Pathol. Oncol. Res. 30:1611993. doi: 10.3389/pore.2024.1611993
Received
07 October 2024
Accepted
17 December 2024
Published
07 January 2025
Volume
30 - 2024
Edited by
Zsolt Orosz, Nuffield Orthopaedic Centre, United Kingdom
Updates
Copyright
© 2025 Lippai, Papp, Szuhai, Sápi, Dezső and Sápi.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zoltán Sápi, sapi.zoltan.dr@gmail.com
†These authors have contributed equally to this work
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.