Abstract
Background: Papillary thyroid carcinoma (PTC) is the most common type of thyroid cancer. The RET gene rearrangements CCDC6::RET and NCOA4::RET are the most common RET gene rearrangements in PTC patients. Different RET::PTC rearrangements are associated with different PTC phenotypes.
Methods: Eighty-three formalin-fixed paraffin-embedded (FFPE) PTC samples were examined. The prevalence and expression levels of CCDC6::RET and NCOA4::RET were determined using semi-quantitative polymerase chain reaction (qRT-PCR). The association of these rearrangements with clinicopathological data was investigated.
Results: The presence of CCDC6::RET rearrangement was significantly associated with the classic subtype and absence of angio/lymphatic invasion (p < 0.05). While NCOA4::RET was associated with the tall-cell subtype, and presence of angio/lymphatic invasion and lymph node metastasis (p < 0.05). Multivariate analysis demonstrated that an absence of extrathyroidal extension and extranodal extension were independent predictive factors for CCDC6::RET, whereas the tall-cell subtype, large tumor size, angioinvasion, lymphatic invasion and perineural invasion were independent predictive factors for NCOA4::RET (p < 0.05). However, the mRNA expression level of CCDC6::RET and of NCOA4::RET were not significantly associated with clinicopathological data.
Conclusion: CCDC6::RET was correlated with an innocent PTC subtype and characteristics, but NCOA4::RET correlated with an aggressive phenotype of PTC. Therefore, these RET rearrangements strongly associated with clinicopathological phenotypes and can be used as predictive markers in PTC patients.
Introduction
Papillary thyroid carcinoma (PTC) is the most common form of well-differentiated thyroid cancer, originating from thyroid follicular cells. PTC is also the most prevalent form of thyroid cancer overall, contributing to 75%–85% of thyroid-cancer cases (1). Although its incidence is rapidly increasing, PTC has the best overall prognosis of any type of thyroid cancer.
BRAF and RAS genes, as well as rearrangements of the RET gene are strongly associated with PTC, respectively. These mutations are associated with mitogen-activated protein kinase (MAPK) and phosphatidylinositol-3 kinase (PI3K)/AKT signaling pathways that affect proliferation and differentiation in cancer cells. RET fusions/rearrangements, are the most frequent molecular alterations reported in PTC, being present in about 10%–40% of cases worldwide, but in 45%–60% of cases involving adolescents and children with sporadic PTC (2–5). The major fusion partner genes in such rearrangements are CCDC6 and NCOA4 (also known as ELE1). These two fusions account for approximately 90% of such cases (6).
The RET (rearranged during transfection) gene is a proto-oncogene located on the long arm of chromosome 10 (10q11.2), and consists of three parts; the extracellular segment, transmembrane segment and an intracellular segment (7); the intracellular segment is an important part that causing of RET::PTC rearrangements. The fusion of tyrosine kinase domain with the 5′region of another gene such as CCDC6 and ELE1, generates chimeric products collectively named RET::PTCs. These result in ligand-independent dimerization and constitutive activation of these chimeric proteins triggers RAS which activates RAF kinases, and the downstream signaling cascade, causing proliferation and differentiation in cancer cells.
CCDC6::RET is formed by fusion with the CCDC6 (also designated H4) gene and NCOA4::RET by fusion with the NCOA4 (also designated ELE1, RFG or ARA70). These rearrangements are associated with papillary thyroid carcinoma cases with different characteristics; CCDC6::RET is more associated with sporadic thyroid cancer (8, 9) and with older patients and classic subtypes of papillary carcinoma (8–10). The NCOA4::RET rearrangement correlates with a more aggressive phenotype and a more advanced stage of PTC. Those fusion genes are also strongly associated with short radiation latency and solid subtypes of PTC (8, 9, 11–13). RET::PTC-mediated signaling depends on a specific RET tyrosine residue, Y1062 (numbering refers to the full-length RET protein). Both types of RET fusion have comparable levels of Y1062 phosphorylation but, MAPK phosphorylation is higher in NCOA4::RET than in CCDC6::RET (14). Therefore, the two rearrangements have the same enzymatic activity and autophosphorylation levels, but activation of MAPK is greater by NCOA4::RET than CCDC6::RET.
Although the global prevalence of major RET::PTC fusions in PTC patients has been reported, but no data are available specifically for Thailand. Thus, this study aimed to determine the prevalence of CCDC6::RET and NCOA4::RET fusions in Thai papillary thyroid carcinoma patients. Prevalence and expression levels of CCDC6::RET and NCOA4::RET rearrangements in PTC patients were determined using semi-quantitative polymerase chain reaction (qRT-PCR). The association of these RET::PTC rearrangements with clinicopathological data was also investigated to test the hypothesis that CCDC6::RET and NCOA4::RET fusion genes are differently associated with aggressive characteristics in PTC.
Material and methods
Study population
The study was approved by the Ethics Committee for Human Research, Khon Kaen University (HE641011). Formal written informed consent was not required with a waiver by the appropriate IRB and/or national research ethics committee. Formalin-fixed paraffin-embedded (FFPE) tissue of patients (n = 83) who had been diagnosed with papillary thyroid carcinoma by pathologists at Pathology services, Department of Pathology in the years 2010–2014 were investigated. All cases were included for this study by clinical data from medical records at Srinagarind hospital, Khon Kaen, Thailand, including sex, age, clinicopathological information including size of tumor, subtype of PTC, angioinvasion, lymphatic invasion, perineural invasion, extrathyroidal extension, extranodal extension and lymph node metastasis. Data relating to lymph node metastasis and extranodal extension was missing for some patients. BRAF and KRAS mutations were not included in clinicopathological data according to genetic alterations were not performed in our routine service. As only morphologic features were investigated for pathological diagnosis.
FFPE slides preparation
Section of formalin-fixed, paraffin-embedded (FFPE) tissues from normal thyroid (n = 10), benign multinodular goiter (n = 10), PTC (n = 83) were stained with hematoxylin and eosin (H&E) and reviewed by two pathologists (SW & PI) to confirm the diagnoses and subclassify the subtypes of papillary carcinomas (Figure 2A). Paraffin blocks of each tumor were examined by pathologists to identify the extent of tumorous tissue. These were then used for RNA extraction.
RNA extraction and cDNA synthesis
FFPE blocks of all PTC tissues, as well as, benign and normal thyroid, were cut to provide sections of approximately 10 µm in thickness. Ten ribbons from each sample were used for RNA extraction. Tissue deparaffinization was performed using xylene and ethanol (SAV Liquid Production GmbH, Flintsbach am Inn, Germany). RNA was extracted using TRIzol® Reagent (Thermofisher Waltham, Massachusetts, United States) with the addition of 0.2 mL of chloroform per 1 mL of TRIZOL reagent was added. Tissue sections were homogenized in 1 mL of TRIZOL reagent per 50–100 mg of tissue (1 ribbon with 10 µm ≈ 1 mg). Isopropanol per 1 mL TRIZOL reagent was used for precipitating the RNA. The concentration and purity of extracted RNA and protein were measured using a nanodrop spectrophotometer at wavelengths of 260 nm (A260) and 280 nm (A280), respectively. The concentration of RNA 50 μg/mL absorbed light at a wavelength of 260 nm in general. Complementary deoxyribonucleic acid (cDNA) was synthesized from messenger RNA using reverse transcription PCR and the RevertAid First Strand cDNA Synthesis Kit (Thermofisher Waltham, Massachusetts, United States) following the manufacturer’s instructions. For oligo (dT) 18 or gene-specific primed cDNA synthesis, incubated for 60 min at 42°C. For random hexamer primed synthesis, incubated for 5 min at 25°C followed by 60 min at 42°C. Terminated the reaction by heating at 70°C for 5 min. The reverse transcription reaction product could be directly used in PCR applications or stored at −20°C for less than 1 week.
Semi-quantitative polymerase chain reaction (qRT-PCR)
Two specific primers were designed based on the sequences deposited in GenBank for CCDC6::RET and NCOA4::RET (Table 1). The nucleotide positions of those primers were synthesized as positive control. cDNA (50 ng/μL) and 23 μL of RT-PCR master mix [SYBR Green master mix qPCR kit (Thermo Scientific Ltd., United Kingdom)] were used for the qRT-PCR. The reactions were carried out in triplicate. GAPDH mRNA was used as an internal control and a no-template control (NTC) was included each time. The amplification of cDNA was performed using a Thermal kit (Thermo Scientific Ltd., United Kingdom) with the following 40 cycles of amplification conditions in Supplementary Table S1. The cuff off cycle threshold (Ct) value was 37th cycle. Cases with Ct values ≤37 were categorized as positive for the specific rearrangement. Post-PCR fluorescence melting curve analysis was done to ensure that only a single product had been amplified. The CT values of CCDC6::RET and NCOA4::RET rearrangements were calculated, and data were expressed as the fold change of control (normal) using 2-(ΔΔ Ct).
TABLE 1
| Name | Primers | Product size (bp) |
|---|---|---|
| GAPDH | F-GTCTCCTCTGACTTCAACAGCG | 97 |
| R-ACCACCCTGTTGCTGTAGCCAA | ||
| CCDC6::RET | F-ATACTGGAGAACAGGCCATCTG | 207 |
| R-CGTTGCCTTGACCACTTTTCC | ||
| NCOA4::RET | F-CCTTTCAGCGAATGGCTCCT | 136 |
| R- ATTCCCACTTTGGATCCTCCT |
The primer sequences.
Nucleotide positions of the primers are based on the sequences deposited in GenBank: CCDC6::RET (accession no.AF395885.1) and NCOA4::RET (accession no. S71225.1).
Gel electrophoresis and sequencing
PCR products from the cases positive according to qRT-PCR were analyzed by electrophoresis through a 2% gel electrophoresis and visualized by ethidium bromide staining. The anticipated sizes of CCDC6::RET and NCOA4::RET products were 207 and 136 bp respectively. These bands were interpreted by visualization with a gel documentation system. Barcode-Tagged Sequencing (BTSeq ™) (U2Bio, Korea) was also applied for positive cases to obtain sequences of the PCR products.
Statistical analyses
Distributions of all continuous variables were tested for normality using the Shapiro-Wilk test. None was found to be normally distributed, requiring the use of non-parametric tests for further analysis. Comparison of categorical variables was carried out using Chi-square tests. Kruskal-Wallis tests and Mann-Whitney U tests to determine whether medians were different between comparison groups and to test for different expression levels between normal, benign goiter and PTC groups. Simple logistic regression and multiple logistic regression were used for backward stepwise univariate and multivariate analyses to identify independent prognostic factors for PTC patients using categorical data. Simple linear regression and multiple linear regression were used for continuous data. Variable factors with p value equal or less than 0.25 from univariate analysis were included to multivariate analysis to investigate the correlation between RET rearrangement and clinical characteristics.
The Ct values in the qRT-PCR assay of CCDC6::RET and NCOA4::RET levels were calculated, and data were expressed as the fold-change relative to controls using 2(−ΔΔCt). Results were regarded as statistically significant at p-values lower than 0.05. The statistical analyses were done using IBM SPSS statistics software version 28.0 (IBM, Chicago, IL, United States).
Results
Prevalence of RET rearrangements
Eighty-three PTC patients included 59 females and 24 males with an average age of 48.21 years (range 17–79 years). Results from qRT-PCR showed the absence of CCDC6::RET or NCOA4::RET rearrangements in normal tissues (n = 10) and benign (multinodular goiter, n = 10) cases. On the other hand, CCDC6::RET rearrangements were detected in 52 PTC cases (62.7%), and NCOA4::RET rearrangements in 43 cases (51.8%). Both CCDC6::RET and NCOA4::RET rearrangements co-occurred in 25 PTC tissues (30.1%). Thirteen PTCs (15.6%) tested negative for either RET::PTC rearrangement (Figure 1).
FIGURE 1
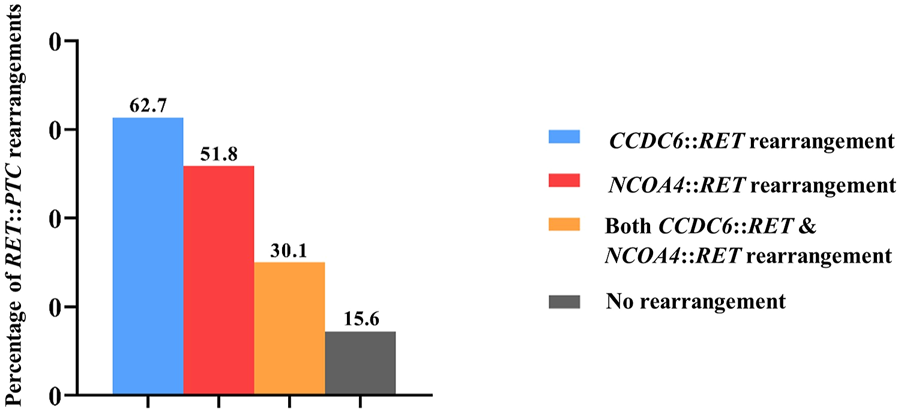
The frequency of RET rearrangements in PTC patients. In detail, CCDC6::RET rearrangements were detected in 62.7% (52/83), NCOA4::RET rearrangements in 51.8% (43/83), both CCDC6::RET and NCOA4::RET rearrangements co-occurred in 30.1% (25/83) and 15.6% (13/83) PTCs were negative for both RET::PTC rearrangements.
The association between RET rearrangements and clinicopathological data in PTC patients
PTCs have been categorized into classic, tall-cell subtypes and follicular variants. A correlation between the histological subtype and the type of RET rearrangement was observed; 71.93% of the classic subtype samples were positive for CCDC6::RET and 42.11% for NCOA4::RET, 43.75% of the tall-cell subtype tissues were positive for CCDC6::RET and 81.25% of them were NCOA4::RET positive. Both the frequency of CCDC6::RET and NCOA4::RET rearrangements were significantly different in those subtypes (OR = 0.27, 95% CI = 0.11–0.71) (Figure 2; Table 2).
FIGURE 2
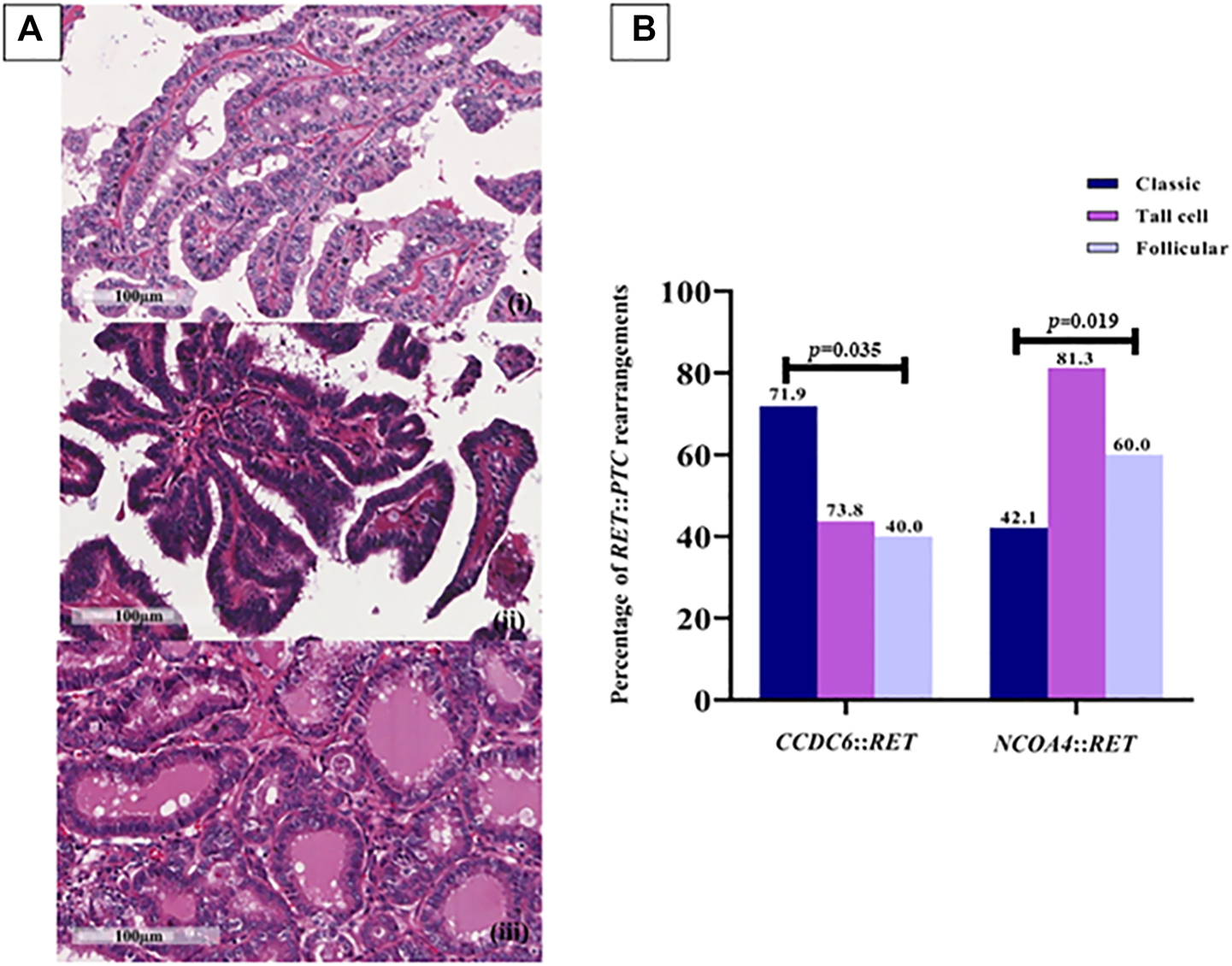
RET rearrangements in each PTC subtype. (A) Papillary thyroid carcinoma subtypes. (i) Classic PTC; consists of papillae with fibrovascular core line by neoplastic cell characterized by increased nuclear size, nuclear overlapping, nuclear features like clear, ground glass appearance and oval shape (400X, scale bar = 100 µm); (ii) Tall cell PTC; abundant eosinophilic cytoplasm and nuclear features characteristic of classic subtype (400X, scale bar = 100 µm); (iii) Follicular PTC; follicular architecture but enlarged nuclei with cytologic features of PTC (400X, scale bar = 100 µm). (B) The prevalence of RET::PTC rearrangements in each PTC subtype; 41 (71.9%) and 24 (42.1%) cases of the classic subtype are positive for CCDC6::RET and NCOA4::RET, respectively, 7 (43.8%) cases of the tall-cell subtype were positive for CCDC6::RET and 13 (81.3%) cases for NCOA4::RET rearrangement, 4 (40%) cases of follicular PTC for CCDC6::RET and 6 (60%) cases for NCOA4::RET. Either the frequency of CCDC6::RET or NCOA4::RET rearrangements are significantly different in those subtypes (p = 0.035, p = 0.019).
TABLE 2
| Variable | Category | N | CCDC6::RET rearrangement (%) | p | NCOA4::RET rearrangement (%) | p | CCDC6::RET and NCOA4::RET rearrangement (%) | p |
|---|---|---|---|---|---|---|---|---|
| Gender | Male | 24 | 15 (62.5) | 0.986 | 14 (58.3) | 0.448 | 9 (37.5) | 0.350 |
| Female | 59 | 37 (62.7) | 29 (49.2) | 16 (27.1) | ||||
| Age | <55 | 53 | 39 (73.6) | 0.006 | 25 (47.2) | 0.261 | 17 (32.1) | 0.606 |
| ≥55 | 30 | 13 (43.3) | 18 (60.0) | 8 (26.7) | ||||
| Tumor size | ≤1 | 7 | 5 (71.4) | 0.311 | 4 (57.1) | 0.501 | 3 (42.9) | 0.746 |
| 1 < T ≤ 2 | 18 | 13 (72.2) | 8 (44.4) | 5 (27.8) | ||||
| 2 < T ≤ 4 | 40 | 26 (65.0) | 19 (47.5) | 13 (32.5) | ||||
| >4 | 18 | 8 (44.4) | 12 (66.7) | 4 (22.2) | ||||
| PTC subtype | Classic | 57 | 41 (71.9) | 0.035 | 24 (42.1) | 0.019 | 14 (24.6) | 0.258 |
| Tall cell | 16 | 7 (43.8) | 13 (81.3) | 7 (43.8) | ||||
| Folliculara | 10 | 4 (40.0) | 6 (60.0) | 4 (40.0) | ||||
| Angioinvasion | Absent | 43 | 33 (76.7) | 0.006 | 15 (34.9) | 0.001 | 13 (30.2) | 0.982 |
| Present | 40 | 19 (47.5) | 28 (70.0) | 12 (30.0) | ||||
| Lymphatic invasion | Absent | 46 | 37 (80.4) | <0.001 | 17 (37.0) | 0.003 | 16 (34.8) | 0.343 |
| Present | 37 | 15 (40.5) | 26 (70.3) | 9 (24.3) | ||||
| Perineural invasion | Absent | 69 | 46 (66.7) | 0.093 | 33 (47.8) | 0.107 | 22 (31.9) | 0.437 |
| Present | 14 | 6 (42.9) | 10 (71.4) | 3 (21.4) | ||||
| Extrathyroidal extension | Absent | 67 | 48 (71.6) | <0.001 | 29 (43.3) | 0.001 | 21 (31.3) | 0.619 |
| Present | 16 | 4 (25.0) | 14 (87.5) | 4 (25.0) | ||||
| Lymph node metastasisb | Absent | 12 | 9 (75.0) | 0.129 | 4 (33.3) | <0.001 | 4 (33.3) | 0.425 |
| Present | 39 | 20 (51.3) | 29 (74.4) | 14 (35.9) | ||||
| Extranodal extensionb | Absent | 31 | 22 (71.0) | 0.030 | 18 (58.1) | 0.013 | 13 (41.9) | 0.180 |
| Present | 19 | 7 (36.8) | 14 (73.7) | 5 (26.3) |
Correlation between RET rearrangements and clinicopathological data using Chi-square analysis.
Bold values mean “statistical significant”.
Categorical variables were compared using Chi-square tests. Neither type of rearrangement was statistically significantly associated with gender. The CCDC6::RET rearrangement was significantly associated with patients younger than 55 years of age (p = 0.006) and classic subtype (p = 0.035). For clinicopathological markers, the CCDC6::RET rearrangement was significantly associated with an absence of angioinvasion (p = 0.006) and of lymphatic invasion extrathyroidal extension and extranodal extension (p= <0.001 and 0.030). NCOA4::RET rearrangements were statistically significantly associated with the tall cell subtype (p = 0.019) and angioinvasion (p = 0.001), lymphatic invasion (p = 0.003), lymph node metastasis (p= <0.001), extrathyroidal extension (p = 0.001) and extranodal extension (p = 0.013). Co-occurrence of CCDC6::RET and NCOA4::RET rearrangements was not significantly associated with clinicopathological data in PTC patients (Table 2).
Following gel electrophoresis and visualization, aliquots of each qRT-PCR product were sent to BTSeq ™ (U2Bio, Korea) for sequencing. The CCDC6::RET and NCOA4::RET products were 207 and 136 bp in length, respectively, as expected (Figure 3). The sequencing results showing the fusion points and sequencing electropherograms are shown in Figure 4.
FIGURE 3
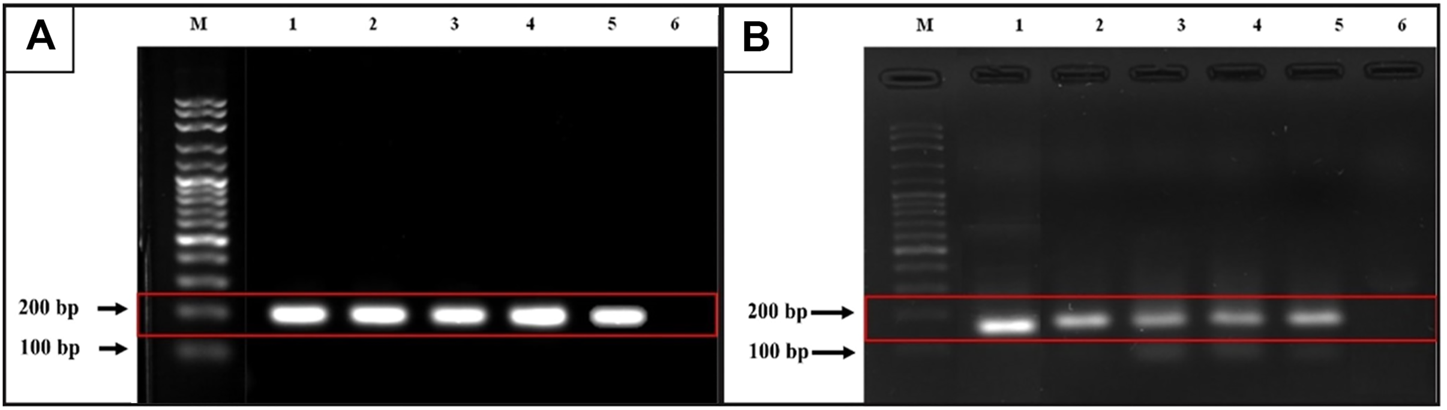
Gel electrophoresis from qRT-PCR products. (A)CCDC6::RET rearrangement (expected band size 207 bp). M = 100 bp ladder, lane 1 is a positive control, lanes 2 and 3 = classic subtype, lane 4 and lane 5 = follicular variant, lane 6 = undetectable case (B)NCOA4::RET rearrangement (expected band size 136 bp). M = 100 bp ladder, lane 1 is a positive control, lane 2 = classic subtype, lane 3 and 4 = tall-cell, lane 5 = follicular variant and lane 6 = undetectable case.
FIGURE 4
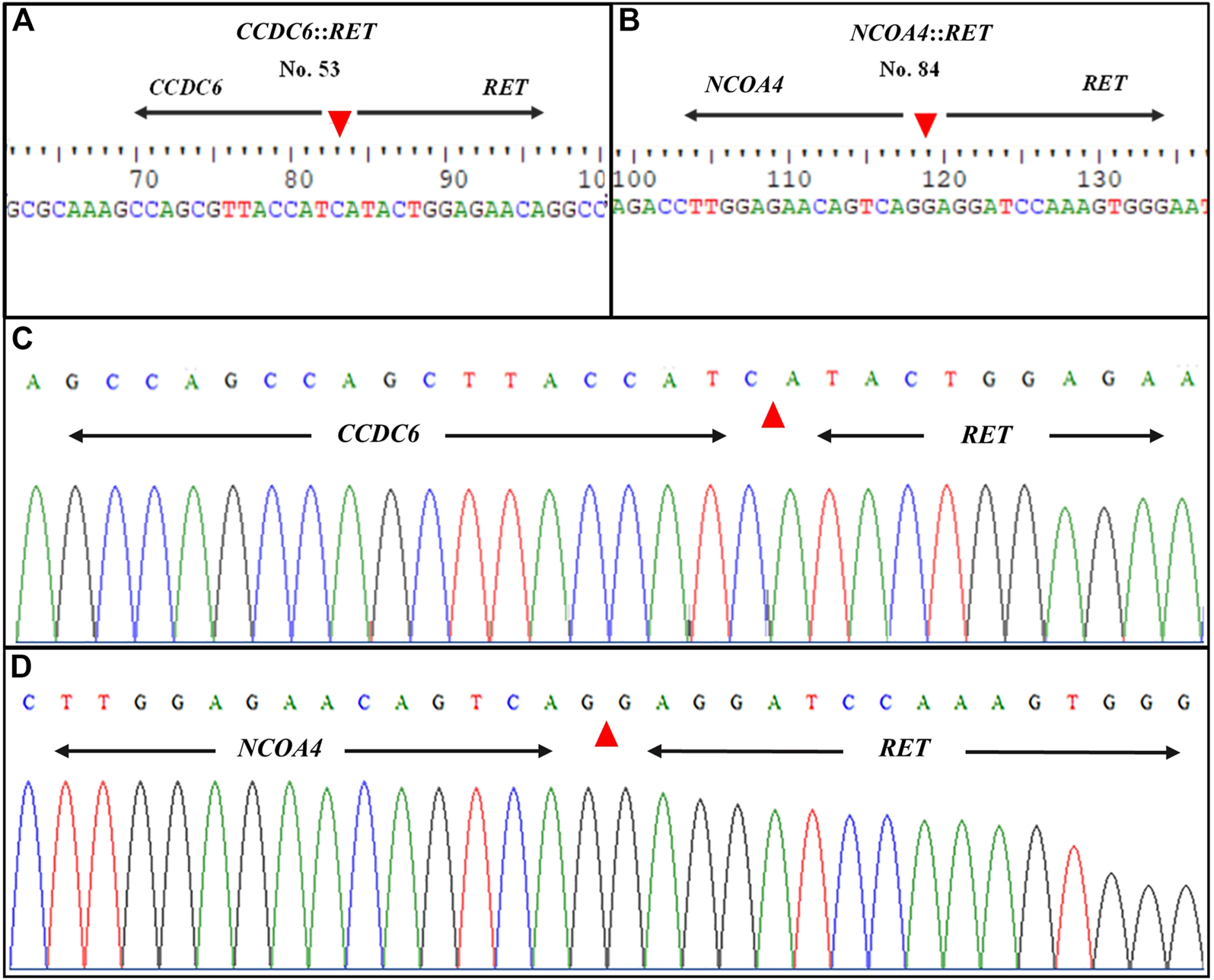
Representative sequence lengths of qRT-PCR products and electropherograms of RET rearrangements. (A)CCDC6::RET; RET fusion with the CCDC6 gene and (B)NCOA4::RET; RET fusion with the NCOA4 gene (C) Showing formed, distinctive single-coloured peaks of DNA electropherogram of CCDC6::RET and (D)NCOA4::RET rearrangements. The patient’s identification number is listed at the top of each panel. Nucleotide sequences around the fusion points of the two different types of RET rearrangement transcripts are shown. Arrowheads indicate the fusion point.
Clinicopathological variables as predictive factors for RET rearrangements
Clinicopathological parameters of the 83 PTC patients were analyzed as dependent and independent predictive factors using simple logistic regression and multiple logistic regression with 95% CI values. Being younger than 55 years was a dependent predictive factor for the CCDC6::RET rearrangement (OR = 0.27, 95% CI = 0.11–0.71; p = 0.007). The absence of angioinvasion (OR = 0.27, 95% CI = 0.11–0.70; p = 0.007), absence of lymphatic invasion (OR = 0.17, 95% CI = 0.06–0.44; p < 0.001), of extrathyroidal extension (OR = 0.13, 95% CI = 0.04–0.46; p = 0.001) and of extranodal extension (OR = 0.24, 95% CI = 0.07–0.80; p = 0.021) were dependent predictive factors for CCDC6::RET. In multivariate analysis, controlling for the influence of other factors, an absence of extrathyroidal extension (OR = 0.25, 95% CI = 0.02–0.84, p = 0.032) and of extranodal extension (OR = 0.16, 95% CI = 0.03–0.80, p = 0.026) were significant independent predictive factors for CCDC6::RET (Table 3).
TABLE 3
| Variable | Category | n (%) | CCDC6::RET rearrangement | n (%) | NCOA4::RET rearrangement | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | ||||||||
| OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p | ||||
| Gender | Male | 15 (62.5) | 1 | 0.986 | 14 (58.3) | 1 | 0.449 | ||||
| Female | 37 (62.7) | 1.01 (0.38–2.69) | 29 (49.2) | 0.69 (0.26–1.80) | |||||||
| Age | <55 | 39 (73.6) | 1 | 0.007 | 1 | 0.159 | 25 (47.2) | 1 | 0.263 | ||
| ≥55 | 13 (43.3) | 0.27 (0.11–0.71) | 0.27 (0.04–1.67) | 18 (60.0) | 1.68 (0.68–4.17) | ||||||
| Tumor size | ≤1 | 5 (71.4) | 1 | 1 | 0.056 | 4 (57.1) | 1 | 1 | |||
| 1 < T ≤ 2 | 13 (72.2) | 1.04 (0.15–7.22) | 0.968 | 8 (44.4) | 0.60 (0.10–3.49) | 0.570 | |||||
| 2 < T ≤ 4 | 26 (65.0) | 0.74 (0.13–4.33) | 0.741 | 19 (47.5) | 1.68 (0.13–3.43) | 0.064 | 1.02 (0.00–1.53) | 0.020 | |||
| >4 | 8 (44.4) | 0.32 (0.05–2.11) | 0.236 | 18.3 (0.93–361.64) | 12 (66.7) | 1.50 (0.25–8.98) | 0.066 | 1.02 (0.00–1.87) | 0.042 | ||
| PTC subtype | Classic | 41 (71.9) | 1 | 24 (43.8) | 1 | 1 | |||||
| Tall cell | 7 (43.8) | 0.30 (0.10–0.95) | 0.041 | 0.75 (0.33–2.58) | 0.07 | 13 (81.3) | 5.96 (1.53–23.24) | 0.010 | 6.06 (2.15–20.49) | 0.032 | |
| Follicular | 4 (40.0) | 1.18 (0.06–1.05) | 0.058 | 1.09 (0.9–3.54) | 0.121 | 6 (60.0) | 2.06 (0.52–8.12) | 0.300 | 0.03 (0.40–5.70) | 0.173 | |
| Angioinvasion | Absent | 33 (76.7) | 1 | 0.007 | 0.48 (0.01–1.34) | 0.086 | 15 (34.9) | 1 | 0.002 | 1 | 0.019 |
| Present | 19 (47.5) | 0.27 (0.11–0.70) | 28 (70.0) | 4.36 (1.73–10.95) | 16.29 (1.57–168.81) | ||||||
| Lymphatic invasion | Absent | 37 (80.4) | 1 | <0.001 | 1 | 0.078 | 17 (37.0) | 1 | 0.003 | 1 | 0.015 |
| Present | 15 (40.5) | 0.17 (0.06–0.44) | 0.25 (0.05–1.17) | 26 (70.3) | 4.03 (1.60–10.17) | 44.11 (2.11–922.55) | |||||
| Perineural invasion | Absent | 46 (66.7) | 1 | 0.101 | 1 | 0.218 | 33 (47.8) | 1 | 0.116 | 1 | 0.039 |
| Present | 6 (42.9) | 0.38 (0.12–1.21) | 0.35 (0.07–1.86) | 10 (71.4) | 2.73 (0.78–9.54) | 24.64 (1.18–515.96) | |||||
| Extrathyroidal extension | Absent | 48 (71.6) | 1 | 0.001 | 1 | 0.032 | 29 (43.3) | 1 | 0.005 | 1 | 0.109 |
| Present | 4 (25.0) | 0.13 (0.04–0.46) | 0.25 (0.02–0.84) | 14 (87.5) | 9.17 (1.93–43.58) | 17.99 (0.53–613.89) | |||||
| Lymph node metastasis | Absent | 9 (75.0) | 1 | 0.157 | 0.89 (0.09–1.2) | 0.123 | 4 (33.3) | 1 | 0.004 | 1 | 0.058 |
| Present | 20 (51.3) | 0.35 (0.08–1.50) | 29 (74.4) | 8.7 (1.96–38.65) | 9.03 (0.93–88.02) | ||||||
| Extranodal extension | Absent | 22 (71.0) | 1 | 0.021 | 1 | 0.026 | 18 (58.1) | 1 | 0.268 | ||
| Present | 7 (36.8) | 0.24 (0.07–0.80) | 0.16 (0.03–0.80) | 14 (73.7) | 2.02 (0.58–7.03) | ||||||
Univariate and multivariate analysis of the clinicopathological factors with RET rearrangements by the simple logistic regression and multiple logistic regression.
Bold values mean “statistical significant”.
Tall-cell PTC (OR = 5.96, 95% CI = 1.53–23.24, p = 0.01), angioinvasion (OR = 4.36, 95% CI = 1.73–10.95; p = 0.002), lymphatic invasion (OR = 4.03, 95% CI = 1.60–10.17; p = 0.003), extrathyroidal extension (OR = 9.17, 95% CI = 1.93–43.58; p = 0.005) and lymph node metastasis (OR = 4.03 95% CI = 1.60–10.17; p = 0.004) were dependent predictive factors of the NCOA4::RET rearrangement. In multivariate analysis, the tall-cell subtype (OR = 6.06, 95% CI = 2.15–20.49, p = 0.032), tumor size 2 < T ≤ 4 cm and large tumor size >4 cm (OR = 1.02, 95% CI = 0.00–1.53, 0.00–0.87; p = 0.020 and 0.042, respectively), perineural invasion (OR = 24.64, 95% CI = 1.18–515.96; p = 0.039) lymphatic invasion (OR = 44.11, 95% CI = 2.11–922.55; p = 0.015), were significant independent predictive factors of NCOA4::RET (Table 3). However, co-occurrence of the two major RET fusions was not significantly associated with the clinicopathological data (Table 4).
TABLE 4
| Variable | Category | n (%) | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|---|
| OR (95% CI) | p | OR (95% CI) | p | |||
| Gender | Male | 9 (37.5) | 1 | 0.352 | ||
| Female | 16 (27.1) | 0.62 (0.23–1.70) | ||||
| Age | <55 | 17 (32.1) | 1 | 0.606 | ||
| ≥55 | 8 (26.7) | 0.77 (0.29–2.08) | ||||
| Tumor size | ≤1 | 3 (42.9) | 1 | |||
| 1 < T ≤ 2 | 5 (27.8) | 0.51 (0.08–3.16) | 0.472 | |||
| 2 < T ≤ 4 | 13 (32.5) | 0.64 (0.12–3.30) | 0.596 | |||
| >4 | 4 (22.2) | 0.38 (0.06–2.46) | 0.310 | |||
| PTC subtype | Classic | 14 (24.6) | 1 | 0.105 | ||
| Tall cell | 7 (43.8) | 2.39 (0.75–7.60) | 0.140 | 1 | ||
| Follicular | 4 (40.0) | 2.05 (0.50–8.32) | 0.316 | 2.71 (0.81–9.04) | ||
| Angioinvasion | Absent | 13 (30.2) | 1 | 0.982 | ||
| Present | 12 (30.0) | 0.99 (0.39–2.53) | ||||
| Lymphatic invasion | Absent | 16 (34.8) | 1 | 0.304 | ||
| Present | 9 (24.3) | 0.60 (0.23–1.58) | ||||
| Perineural invasion | Absent | 22 (31.9) | 1 | 0.441 | ||
| Present | 3 (21.4) | 0.58 (0.15–2.30) | ||||
| Extrathyroidal extension | Absent | 21 (31.3) | 1 | 0.620 | ||
| Present | 4 (25.0) | 0.73 (0.21–2.53) | ||||
| Lymph node metastasis | Absent | 4 (33.3) | 1 | 0.871 | ||
| Present | 14 (35.9) | 1.12 (0.29–4.39) | ||||
| Extranodal extension | Absent | 13 (41.9) | 1 | 0.268 | ||
| Present | 5 (26.3) | 0.49 (0.14–1.72) | ||||
Univariate and multivariate analysis of the clinicopathological factors with both CCDC6::RET and NCOA4::RET rearrangements group by the simple logistic regression and multiple logistic.
Determination of mRNA expression levels of RET rearrangements in thyroid tissues
The mRNA expression levels of CCDC6::RET and NCOA4::RET rearrangements in PTC compared to benign (multinodular goiter) and normal thyroid tissue are shown in Figure 5. The median mRNA expression levels of CCDC6::RET transcripts were 0.61 (0.14–1.34), 0.76 (0.36–1.89) and 1.09 (0.20–328.60) in normal, benign and PTC tissues, respectively. The median mRNA expression levels of NCOA4::RET rearrangement were 0.66 (0.08–2.19), 0.76 (0.36–1.89) and 3.59 (0.69–162.38) in normal, benign and PTC tissues, respectively. The relative CCDC6::RET mRNA expression levels in PTC (1.79) and benign tissues (1.25) were significantly higher than in normal tissues (p = 0.003). The relative mRNA expression levels of NCOA4::RET transcripts were significantly higher in PTC tissues (5.44) than in benign tissues (1.15) or normal tissues (p < 0.001 in both cases) (Figure 5).
FIGURE 5
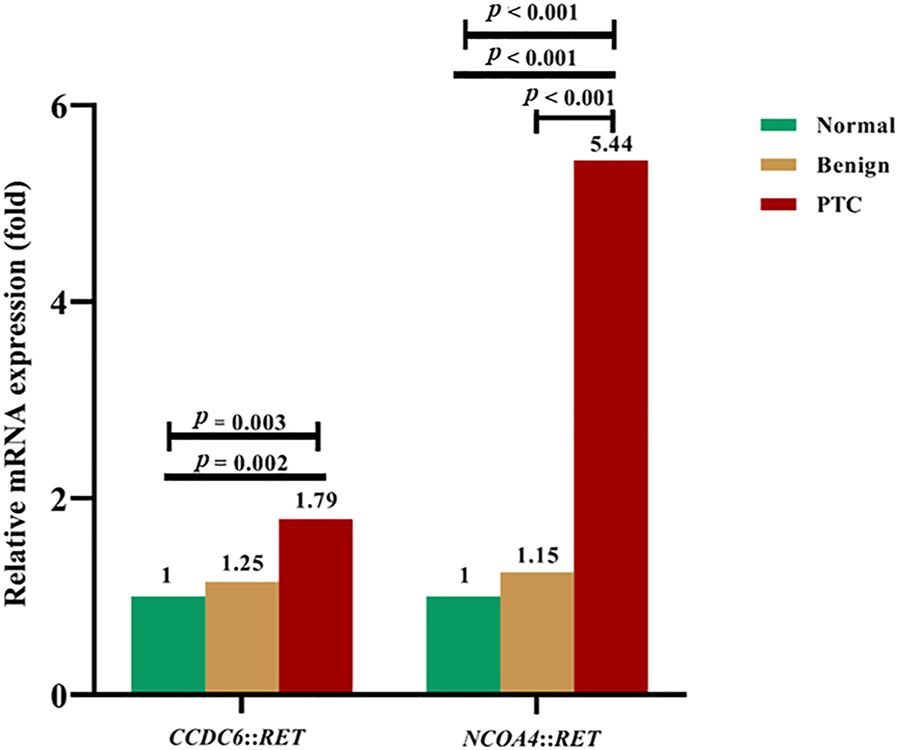
The relative fold-change of mRNA expression levels in RET rearrangements transcripts in normal, benign and PTC tissues. For CCDC6::RET, showing the differentiation of mRNA expression among PTC, benign compared to normal tissue (p = 0.003) by Kruskal-Wallis test. Expression of mRNA in PTC is greater than in normal thyroid tissue (p = 0.002) by Mann Whitney U test. The NCOA4::RET mRNA expression, showing the differentiation of mRNA expression among PTC, benign and normal tissues (p < 0.001), normal tissue compared to PTC (p < 0.001), and benign tissue compared to PTC (p < 0.001).
The associations between clinicopathological factors and relative mRNA expression levels of RET rearrangements in PTC patients
Associations between clinicopathological factors and mRNA expression levels of RET rearrangements were performed using Mann-Whitney U tests and Kruskal-Wallis tests. No statistically significant association with clinicopathological factors was found (Table 5).
TABLE 5
| Variable | Category | n (%) | CCDC6::RET mRNA expression | n (%) | NCOA4::RET mRNA expression | ||
|---|---|---|---|---|---|---|---|
| Median | p | Median | p | ||||
| Gender | Male | 15 (62.5) | 1.00 (0.20–17.23) | 0.762 | 14 (58.3) | 6.13 (1.00–162.38) | 0.734 |
| Female | 37 (62.7) | 1.12 (0.48–328.60) | 29 (49.2) | 3.79 (0.69–95.52) | |||
| Age | <55 | 39 (73.6) | 1.31 (0.20–328.60) | 0.485 | 25 (47.2) | 3.38 (0.69–95.52) | 0.722 |
| ≥55 | 13 (43.3) | 1.06 (0.55–2.63) | 18 (60.0) | 3.88 (0.97–162.38) | |||
| Tumor size | ≤1 | 5 (71.4) | 1.67 (0.49–17.23) | 0.100 | 4 (57.1) | 1.92 (0.69–35.59) | 0.508 |
| 1 < T ≤ 2 | 13 (72.2) | 1.00 (0.64–328.60) | 8 (44.4) | 2.94 (0.97–162.38) | |||
| 2 < T ≤ 4 | 26 (65.0) | 1.06 (0.20–61.12) | 19 (47.5) | 4.88 (0.98–62.24) | |||
| >4 | 8 (44.4) | 0.90 (0.55–1.94) | 12 (66.7) | 5.19 (1.32–93.92) | |||
| PTC subtype | Classic | 41 (71.9) | 1.94 (0.20–328.60) | 0.761 | 24 (42.1) | 3.37 (0.69–52.19) | 0.294 |
| Tall cell | 7 (43.8) | 1.63 (0.64–17.23) | 13 (81.3) | 6.62 (0.69–162.38) | |||
| Follicular | 4 (40.0) | 0.80 (0.49–26.28) | 6 (60.0) | 3.18 (1.56–52.19) | |||
| Angioinvasion | Absent | 33 (76.7) | 1.31 (0.53–328.60) | 0.091 | 15 (34.9) | 3.79 (0.69–62.24) | 0.961 |
| Present | 19 (47.5) | 0.87 (0.20–18.48) | 28 (70.0) | 5.55 (0.69–162.38) | |||
| Lymphatic invasion | Absent | 37 (80.4) | 1.12 (0.20–328.60) | 0.770 | 17 (37.0) | 3.79 (0.97–95.52) | 0.377 |
| Present | 15 (40.5) | 1.04 (0.48–61.12) | 26 (70.3) | 2.86 (0.69–162.38) | |||
| Perineural invasion | Absent | 46 (66.7) | 1.32 (0.20–328.60) | 0.057 | 33 (47.8) | 3.38 (0.69–162.38) | 0.501 |
| Present | 6 (42.9) | 0.66 (0.49–8.07) | 10 (71.4) | 4.56 (1.06–93.92) | |||
| Extrathyroidal extension | Absent | 48 (71.6) | 1.03 (0.20–328.60) | 0.548 | 29 (43.3) | 2.94 (0.69–95.52) | 0.088 |
| Present | 4 (25.0) | 1.50 (1.04–2.63) | 14 (87.5) | 5.55 (1.36–162.38) | |||
| Lymph node metastasis | Absent | 9 (75.0) | 0.92 (0.70–8.07) | 0.814 | 4 (33.3) | 5.39 (0.69–7.37) | 0.934 |
| Present | 20 (51.3) | 1.18 (0.20–61.12) | 29 (74.4) | 3.18 (0.69–162.38) | |||
| Extranodal extension | Absent | 22 (71.0) | 1.02 (0.20–61.12) | 0.415 | 18 (58.1) | 3.88 (0.69–93.92) | 0.985 |
| Present | 7 (36.8) | 0.73 (0.48–4.29) | 14 (73.7) | 2.86 (0.97–162.38) | |||
Mann-Whitney U tests and Kruskal-Wallis test of the predictive marker with relative RET mRNA expression levels in papillary thyroid carcinoma.
In univariate analysis, the clinicopathological factors were not dependent predictive factors for CCDC6::RET and NCOA4::RET mRNA expression levels (Tables 6, 7).
TABLE 6
| Variable | Category | n (%) | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|---|
| Mean diff (95% CI) | p | Mean diff (95% CI) | p | |||
| Gender | Male | 15 (62.5) | 1 | 0.416 | ||
| Female | 37 (62.7) | 11.57 (−16.77–39.92) | ||||
| Age | <55 | 39 (73.6) | 1 | 0.382 | ||
| ≥55 | 13 (43.3) | −13.02 (−42.62–16.60) | ||||
| Tumor size | ≤1 | 5 (71.4) | 1 | 1 | ||
| 1 < T ≤ 2 | 13 (72.2) | 28.87 (−19.26–76.99) | 0.234 | −6.90 (−15.49–1.70) | 0.111 | |
| 2 < T ≤ 4 | 26 (65.0) | 0.37 (−44.29–45.02) | 0.987 | |||
| >4 | 8 (44.4) | −3.12 (−55.26–49.01) | 0.905 | |||
| PTC subtype | Classic | 41 (71.9) | 1 | |||
| Tall cell | 7 (43.8) | −9.23 (−47.66–29.20) | 0.632 | |||
| Follicular | 4 (40.0) | −5.68 (−54.91–43.54) | 0.817 | |||
| Angioinvasion | Absent | 33 (76.7) | 1 | 0.295 | 1 | |
| Present | 19 (47.5) | −13.99 (−40.54–12.57) | ||||
| Lymphatic invasion | Absent | 37 (80.4) | 1 | 0.659 | ||
| Present | 15 (40.5) | −6.30 (−34.78–22.18) | ||||
| Perineural invasion | Absent | 46 (66.7) | 1 | 0.607 | ||
| Present | 6 (42.9) | −10.40 (−50.76–29.96) | ||||
| Extrathyroidal extension | Absent | 48 (71.6) | 1 | 0.674 | ||
| Present | 4 (25.0) | −10.22 (−58.65–38.22) | ||||
| Lymph node metastasis | Absent | 9 (75.0) | 1 | 0.532 | ||
| Present | 20 (51.3) | 2.88 (−6.46–12.21) | ||||
| Extranodal extension | Absent | 22 (71.0) | 1 | 0.515 | ||
| Present | 7 (36.8) | −3.24 (−13.33–6.84) | ||||
Univariate analysis and multivariate analysis of the clinicopathological factors with relative CCDC6::RET mRNA expression level by simple linear regression and multiple linear regression.
TABLE 7
| Variable | Category | n (%) | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|---|
| Mean diff (95% CI) | p | Mean diff (95% CI) | p | |||
| Gender | Male | 14 (58.3) | 1 | 0.363 | ||
| Female | 29 (49.2) | −9.55 (−30.49–11.40) | ||||
| Age | <55 | 25 (47.2) | 1 | 0.356 | ||
| ≥55 | 18 (60.0) | 9.10 (−10.59–28.79) | ||||
| Tumor size | ≤1 | 4 (57.1) | 1 | |||
| 1 < T ≤ 2 | 8 (44.4) | −10.76 (−51.46–29.94) | 0.596 | |||
| 2 < T ≤ 4 | 19 (47.5) | 0.73 (−35.68–37.13) | 0.968 | |||
| >4 | 12 (66.7) | −3.62 (−41.99–34.76) | 0.850 | |||
| PTC subtype | Classic | 24 (42.1) | 1 | 0.051 | ||
| Tall cell | 13 (81.3) | 21.60 (0.02–43.18) | 0.050 | 1 | ||
| Follicular | 6 (60.0) | 9.74 (−18.95–38.43) | 0.497 | 20.5 (0.23–40.77) | ||
| Angioinvasion | Absent | 15 (34.9) | 1 | 0.374 | ||
| Present | 28 (70.0) | 9.03 (−11.26–29.32) | ||||
| Lymphatic invasion | Absent | 17 (37.0) | 1 | 0.808 | ||
| Present | 26 (70.3) | 2.43 (−17.60–22.45) | ||||
| Perineural invasion | Absent | 33 (47.8) | 1 | 0.829 | ||
| Present | 10 (71.4) | 2.53 (−20.97–26.03) | ||||
| Extrathyroidal extension | Absent | 29 (43.3) | 1 | 0.123 | 1 | 0.082 |
| Present | 14 (87.5) | 15.74 (−4.46–35.94) | 18.05 (−2.37–38.48) | |||
| Lymph node metastasis | Absent | 4 (33.3) | 1 | 0.530 | ||
| Present | 29 (74.4) | 11.12 (−24.6–46.85) | ||||
| Extranodal extension | Absent | 18 (58.1) | 1 | 0.558 | ||
| Present | 14 (73.7) | 6.86 (−16.75–30.47) | ||||
Univariate analysis and multivariate analysis of the clinicopathological factors with relative NCOA4::RET mRNA expression level by simple linear regression and multiple linear regression.
Discussion
Papillary thyroid cancer (PTC) is the most common form of thyroid cancer. It is also rapidly increasing in incidence worldwide. This cancer has quite a good prognosis, with less than 2% mortality at 5 years. However, more than 25% of PTC patients suffer a recurrence in the long term (15).
Rearrangements of BRAF, RAS, P53 and RET are all strongly associated with thyroid cancer. For RET rearrangements, RET::PTCs are the most frequent molecular alterations reported in PTC (2–5). Several fusion partner genes of RET in such rearrangements have been reported, but the major ones are CCDC6and NCOA4 (accounting for more than 90%) (6).
We found that the prevalence of CCDC6::RET, NCOA4::RET rearrangements and co-occurrence of both in Thailand were 62.65%, 51.81%, and 30.12%, respectively. These rearrangements were not found in normal thyroid, or in benign tumors. Similarly, high frequencies of RET::PTC rearrangements have been reported from Taiwan (55%), New Caledonia (70%) and Australia (85%) (16, 17). On the other hand, studies from Korea (6.5%) and Japan (30%) showed low frequencies of RET::PTC rearrangements in PTC (18, 19). This heterogeneity in RET::PTC rearrangements may be due to ethnicity, different geographical location and/or differences in environmental exposure. The literature has reported that frequency of RET rearrangements in papillary carcinomas ranges from 0% to 87% (20, 21). These differences are partly explained by pre-selection of the analyzed cohort, geographical diversity and different sensitivities of the assays used for detection (20, 22). Naito et al. (23) reported that the prevalence of RAS gene mutations in Thai PTC patients was higher than in Japan. Being affected the RET rearrangements according to RET rearrangements/mutations and RAS family mutations are almost always mutually exclusive (23, 24).
This suggests that an existence of a specific environmental agent in northeast Thailand, presumably related to the high level of nitrates in the diet, particularly high salt-preserved fish; Pla-ra, Pla-som and also vegetables, water promotes the progression of thyroid papillary carcinomas (23). Apart from the forementioned majority factors, other putative carcinogens, for example, caffeine, ethanol, hypoxia are able to induce DNA double-strand breaks and generate RET/PTC rearrangements (26–28). As reported previously, the frequency of CCDC6::RET in sporadic thyroid cancer is higher than that of NCOA4::RET (8, 9). Moreover, our study revealed no major RET rearrangements in benign tumors, similar to previous studies (13, 21).
CCDC6::RET rearrangement was significantly associated with the classic histological subtype (41/57) (p = 0.035), a result in agreement with previous studies (8–10, 29). Similarly, Guerra et al (4) reported that CCDC6::RET was the dominant type within sporadic carcinomas and was strongly related to the classic subtype (4). NCOA4::RET rearrangement was significantly associated with the tall cell subtype (13/16) (p = 0.019). In term of clinicopathological markers, the NCOA4::RET rearrangement was significantly associated with angioinvasion (p = 0.001), lymphatic invasion (p = 0.003), extrathyroidal extension (p = 0.001), lymph node metastasis (p < 0.001) and extranodal extension (p = 0.013), respectively. This is in agreement with Sugg et al (12) and Mochizuki et al (30) who reported that NCOA4::RET may cause de-differentiation and more aggressive behavior in papillary thyroid carcinoma (12, 30). Certainly, NCOA4::RET correlates with a more aggressive phenotype and a more advanced stage of PTC (8, 9, 11–13). Galuppini et al (31), Khan et al (18) and Rogounovitch et al (32) reported that an NCOA4::RET rearrangement was associated with lymph node metastasis (18, 31, 32).
Univariate analysis revealed CCDC6::RET mRNA expression levels in patients with small tumor size (1 < T ≤ 2 cm) and the classic subtype was not significant that in contrast to with Rhoden et al. (21). Moreover, the other clinicopathological markers, such as extrathyroidal extension or metastases were not correlated with CCDC6::RET, in agreement with Tallini et al (33).
High expression levels of NCOA4::RET were found, but not significant in patients with the tall-cell subtype, extrathyroidal extension, lymph node metastasis and extranodal extension. Our findings are similar to those of from Powell et al (11) that showed the NCOA4::RET rearrangement was more frequent in tall-cell tumors: its expression in transgenic mice generated solid tumor subtypes of thyroid cancer with more aggressive and metastatic behavior (11). Martínez et al (34) reported that high detection or expression of BRAFV600E and low expression or absence of that CCDC6::RET rearrangement were associated with extrathyroidal extension. In contrast, a high activity of RET (presence of CCDC6::RET rearrangement or high expression) and low activity of BRAF (absence of BRAFV600E or its low expression) were associated with lymph node metastasis. It has been suggested that BRAFV600E and the CCDC6::RET rearrangement can predict aggressive PTC (34). The literature suggests that RET::PTC rearrangements, and especially NCOA4::RET, can be induced by radiation (23, 35). In such cases, Nikiforov et al (8) and Rabes HM et al (9) reported that in radiation-induced PTC, NCOA4::RET has an equal or even higher prevalence than CCDC6::RET fusions (8, 9). Radiation exposure is often associated with NCOA4::RET and is more common in children 4 years of age or younger. In addition, the latest WHO classification 2022 emphasizes that the telomerase reverse transcriptase (TERT) gene mutation in PTC patients correlates with an increased risk of distant metastases and poor prognosis. Moreover, the gain of chromosome 1q in the PLEKHS1 promoter (PLEKSH1p) may cause a poorer outcome in PTCs (36). Therefore, further investigation is required into more data on history of recurrence, radiation history, gene aberration, tumor focality among other aspects to enhance our understanding of RET::PTC rearrangements.
In our study, tumor tissues from 83 PTC patients were investigated to determine the prevalence of CCDC6::RET and NCOA4::RET rearrangements in Thailand and to explore the association between the mRNA expression of CCDC6::RET and NCOA4::RET rearrangements and clinicopathology. The CCDC6::RET rearrangement was significantly associated with less aggressive histological subtypes and absence of certain clinicopathological markers. Moreover, the absence of extrathyroidal and extranodal extension were independent predictive factors for CCDC6::RET.
In contrast, large tumor size, more aggressive types like the tall-cell type, angioinvasion, perineural invasion or extrathyroidal extension were independent predictive factors of NCOA4::RET. Briefly, CCDC6::RET correlated with less aggressive, but NCOA4::RET correlated with more aggressive phenotypes of PTC. In conclusion, the two rearrangements that we studied were strongly associated with particular clinicopathological phenotypes, particularly histological subtype.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The study was approved by the Ethics Committee for Human Research, Khon Kaen University (HE641011). Formal written informed consent was not required with a waiver by the appropriate IRB and/or national research ethics committee.
Author contributions
TK: conceptualization, data curation, formal analysis, investigation, methodology, resources, software, validation and writing—original draft, writing—review and editing, and approval of the final manuscript. SW: methodology, visualization, writing—review and editing, and approval of the final manuscript. YC: validation, writing—review and editing, and approval of the final manuscript. PI: methodology, visualization, writing—review and editing, and approval of the final manuscript. RD: formal analysis, funding acquisition, methodology, project administration, resources, supervision, validation, visualization, conceptualization, writing—original draft, writing—review and editing, and approval of the final manuscript.
Funding
The study was supported by a postgraduate study support grant of Faculty of Medicine, Khon Kaen University and Invitation Research Fund from Faculty of Medicine No. IN64147 (date of approval 18 January 2021), Khon Kaen University, Thailand.
Acknowledgments
We would like to extend acknowledgment to Srinagarind Hospital for providing the specimens. We would like to acknowledge Prof. David Blair for editing the manuscript via Publication Clinic KKU, Thailand. We thank Mr. Suwit Balthaisong for preparation the slides from FFPE blocks that were used in this work. We thank Mr. Manop Sripa for molecular technique suggestions. We also thank Miss Piyanan Suparattanagool as the statistical expertise consultants.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.por-journal.com/articles/10.3389/pore.2023.1611138/full#supplementary-material
References
1.
Fagin JA Wells SA Jr . Biologic and clinical perspectives on thyroid cancer. N Engl J Med (2016) 375(11):1054–67. 10.1056/NEJMra1501993
2.
Liu RT Chou FF Wang CH Lin CL Chao FP Chung JC et al Low prevalence of RET rearrangements (RET/PTC1, RET/PTC2, RET/PTC3, and ELKS-RET) in sporadic papillary thyroid carcinomas in taiwan Chinese. Thyroid (2005) 15(4):326–35. 10.1089/thy.2005.15.326
3.
Fenton CL Lukes Y Nicholson D Dinauer CA Francis GL Tuttle RM . The ret/PTC mutations are common in sporadic papillary thyroid carcinoma of children and young adults. J Clin Endocrinol Metab (2000) 85(3):1170–5. 10.1210/jcem.85.3.6472
4.
Guerra A Rosaria Sapio M Marotta V Campanile E Ilaria Moretti M Deandrea M et al Prevalence of RET/PTC rearrangement in benign and malignant thyroid nodules and its clinical application. Endocr J (2011) 58(1):31–8. 10.1507/endocrj.k10e-260
5.
Liu S Gao A Zhang B Zhang Z Zhao Y Chen P et al Assessment of molecular testing in fine-needle aspiration biopsy samples: An experience in a Chinese population. Exp Mol Pathol (2014) 97(2):292–7. 10.1016/j.yexmp.2014.08.005
6.
Staubitz JI Musholt TJ Schad A Springer E Lang H Rajalingam K et al ANKRD26-RET - a novel gene fusion involving RET in papillary thyroid carcinoma. Cancer Genet (2019) 238:10–7. 10.1016/j.cancergen.2019.07.002
7.
Santoro M Carlomagno F . Central role of RET in thyroid cancer. Cold Spring Harb Perspect Biol (2013) 5(12):a009233. 10.1101/cshperspect.a009233
8.
Nikiforov YE Rowland JM Bove KE Monforte-Munoz H Fagin JA . Distinct pattern of ret oncogene rearrangements in morphological variants of radiation-induced and sporadic thyroid papillary carcinomas in children. Cancer Res (1997) 57(9):1690–4.
9.
Rabes HM Demidchik EP Sidorow JD Lengfelder E Beimfohr C Hoelzel D et al Pattern of radiation-induced RET and NTRK1 rearrangements in 191 post-chernobyl papillary thyroid carcinomas: Biological, phenotypic, and clinical implications. Clin Cancer Res (2000) 6(3):1093–103.
10.
Smida J Salassidis K Hieber L Zitzelsberger H Kellerer AM Demidchik EP et al Distinct frequency of ret rearrangements in papillary thyroid carcinomas of children and adults from Belarus. Int J Cancer (1999) 80(1):32–8. 10.1002/(sici)1097-0215(19990105)80:1<32:aid-ijc7>3.0.co;2-l
11.
Powell DJ Russell J Nibu K Li G Rhee E Liao M et al The RET/PTC3 oncogene: Metastatic solid-type papillary carcinomas in murine thyroids. Cancer Res (1998) 58(23):5523–8.
12.
Sugg SL Ezzat S Rosen IB Freeman JL Asa SL . Distinct multiple RET/PTC gene rearrangements in multifocal papillary thyroid neoplasia. J Clin Endocrinol Metab (1998) 83(11):4116–22. 10.1210/jcem.83.11.5271
13.
Thomas GA Bunnell H Cook HA Williams ED Nerovnya A Cherstvoy ED et al High prevalence of RET/PTC rearrangements in Ukrainian and belarussian post-chernobyl thyroid papillary carcinomas: A strong correlation between RETPTC3 and the solid-follicular variant. J Clin Endocrinol Metab (1999) 84(11):4232–8. 10.1210/jcem.84.11.6129
14.
Basolo F Giannini R Monaco C Melillo RM Carlomagno F Pancrazi M et al Potent mitogenicity of the RET/PTC3 oncogene correlates with its prevalence in tall-cell variant of papillary thyroid carcinoma. Am J Pathol (2002) 160(1):247–54. 10.1016/S0002-9440(10)64368-4
15.
Abdullah MI Junit SM Ng KL Jayapalan JJ Karikalan B Hashim OH . Papillary thyroid cancer: Genetic alterations and molecular biomarker investigations. Int J Med Sci (2019) 16(3):450–60. 10.7150/ijms.29935
16.
Lee CH Hsu LS Chi CW Chen GD Yang AH Chen JY . High frequency of rearrangement of the RET protooncogene (RET/PTC) in Chinese papillary thyroid carcinomas. J Clin Endocrinol Metab (1998) 83(5):1629–32. 10.1210/jcem.83.5.4774
17.
Chua EL Wu WM Tran KT Mccarthy SW Lauer CS Dubourdieu D et al Prevalence and distribution of RET/PTC 1, 2, and 3 in papillary thyroid carcinoma in New Caledonia and Australia. J Clin Endocrinol Metab (2000) 85(8):2733–9. 10.1210/jcem.85.8.6722
18.
Khan MS Qadri Q Makhdoomi MJ Wani MA Malik AA Niyaz M et al RET/PTC gene rearrangements in thyroid carcinogenesis: Assessment and clinico-pathological correlations. Pathol Oncol Res (2020) 26(1):507–13. 10.1007/s12253-018-0540-3
19.
Chung JH Hahm JR Min YK Lee MS Lee MK Kim KW et al Detection of RET/PTC oncogene rearrangements in Korean papillary thyroid carcinomas. Thyroid (1999) 9(12):1237–43. 10.1089/thy.1999.9.1237
20.
Zhu Z Ciampi R Nikiforova MN Gandhi M Nikiforov YE . Prevalence of RET/PTC rearrangements in thyroid papillary carcinomas: Effects of the detection methods and genetic heterogeneity. J Clin Endocrinol Metab (2006) 91(9):3603–10. 10.1210/jc.2006-1006
21.
Rhoden KJ Johnson C Brandao G Howe JG Smith BR Tallini G . Real-time quantitative RT-PCR identifies distinct c-RET, RET/PTC1 and RET/PTC3 expression patterns in papillary thyroid carcinoma. Lab Invest (2004) 84(12):1557–70. 10.1038/labinvest.3700198
22.
Musholt TJ Staubitz JI Antonio Cámara RJ Musholt PB Humberg D Springer E et al Detection of RET rearrangements in papillary thyroid carcinoma using RT-PCR and FISH techniques - a molecular and clinical analysis. Eur J Surg Oncol (2019) 45(6):1018–24. 10.1016/j.ejso.2018.11.009
23.
Naito H Pairojkul C Kitahori Y Yane K Miyahara H Konishi N et al Different ras gene mutational frequencies in thyroid papillary carcinomas in Japan and Thailand. Cancer Lett (1998) 131(2):171–5. 10.1016/s0304-3835(98)00149-9
24.
Sherman SI Clary DO Elisei R Schlumberger MJ Cohen EW Schöffski P et al Correlative analyses of RET and RAS mutations in a phase 3 trial of cabozantinib in patients with progressive, metastatic medullary thyroid cancer. Cancer (2016) 122(24):3856–64. 10.1002/cncr.30252
25.
Melillo RM Castellone MD Guarino V Falco VD Cirafici AM Salvatore G et al The RET/PTC-RAS-BRAF linear signaling cascade mediates the motile and mitogenic phenotype of thyroid cancer cells. J Clin Invest (2005) 115(4):1068–81. 10.1172/JCI22758
26.
Romei C Elisei R . RET/PTC translocations and clinico-pathological features in human papillary thyroid carcinoma. Front Endocrinol (2012) 3:54. 10.3389/fendo.2012.00054
27.
Gandhi M Dillon LW Pramanik S Nikiforov YE Wang YH . DNA breaks at fragile sites generate oncogenic RET/PTC rearrangements in human thyroid cells. Oncogene (2010) 29(15):2272–80. 10.1038/onc.2009.502
28.
Gandhi M Evdokimova V Nikiforov YE . Mechanisms of chromosomal rearrangements in solid tumors: The model of papillary thyroid carcinoma. Mol Cel Endocrinol (2010) 321(1):36–43. 10.1016/j.mce.2009.09.013
29.
Adeniran AJ Zhu Z Gandhi M Steward DL Fidler JP Giordano TJ et al Correlation between genetic alterations and microscopic features, clinical manifestations, and prognostic characteristics of thyroid papillary carcinomas. Am J Surg Pathol (2006) 30(2):216–22. 10.1097/01.pas.0000176432.73455.1b
30.
Mochizuki K Kondo T Nakazawa T Iwashina M Kawasaki T Nakamura N et al RET rearrangements and BRAF mutation in undifferentiated thyroid carcinomas having papillary carcinoma components: Genetic background of composite UTC. Histopathology (2010) 57(3):444–50. 10.1111/j.1365-2559.2010.03646.x
31.
Galuppini F Vianello F Censi S Barollo S Bertazza L Carducci S et al Differentiated thyroid carcinoma in pediatric age: Genetic and clinical scenario. Front Endocrinol (2019) 10:552. 10.3389/fendo.2019.00552
32.
Rogounovitch TI Mankovskaya SV Fridman MV Leonova TA Kondratovitch VA Konoplya NE et al Major oncogenic drivers and their clinicopathological correlations in sporadic childhood papillary thyroid carcinoma in Belarus. Cancers (2021) 13(13):3374. 10.3390/cancers13133374
33.
Tallini G Santoro M Helie M Carlomagno F Salvatore G Chiappetta G et al RET/PTC oncogene activation defines a subset of papillary thyroid carcinomas lacking evidence of progression to poorly differentiated or undifferentiated tumor phenotypes. Clin Cancer Res Off J Am Assoc Cancer Res (1998) 4(2):287–94.
34.
Martínez JRW Vargas-Salas S Gamboa SU Muñoz E Domínguez JM León A et al The combination of RET, BRAF and demographic data identifies subsets of patients with aggressive papillary thyroid cancer. Horm Cancer (2019) 10(2–3):97–106. 10.1007/s12672-019-0359-8
35.
Fugazzola L Pilotti S Pinchera A Vorontsova TV Mondellini P Bongarzone I et al Oncogenic rearrangements of the RET proto-oncogene in papillary thyroid carcinomas from children exposed to the chernobyl nuclear accident. Cancer Res (1995) 55(23):5617–20.
36.
Baloch ZW Asa SL Barletta JA Ghossein RA Juhlin CC Jung CK et al Overview of the 2022 WHO classification of thyroid neoplasms. Endocr Pathol (2022) 33(1):27–63. 10.1007/s12022-022-09707-3
Summary
Keywords
papillary thyroid carcinoma, RET rearrangements, CCDC6::RET rearrangement, NCOA4::RET rearrangement, gene rearrangements
Citation
Khonrak T, Watcharadetwittaya S, Chamgramol Y, Intarawichian P and Deenonpoe R (2023) RET rearrangements are relevant to histopathologic subtypes and clinicopathological features in Thai papillary thyroid carcinoma patients. Pathol. Oncol. Res. 29:1611138. doi: 10.3389/pore.2023.1611138
Received
21 February 2023
Accepted
18 April 2023
Published
28 April 2023
Volume
29 - 2023
Edited by
Andrea Ladányi, National Institute of Oncology (NIO), Hungary
Updates
Copyright
© 2023 Khonrak, Watcharadetwittaya, Chamgramol, Intarawichian and Deenonpoe.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Raksawan Deenonpoe, raksde@kku.ac.th
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.