Abstract
Despite significant advances in the diagnosis and treatment of esophageal squamous cell carcinoma (ESCC), esophageal cancer is still a heavy social and medical burden due to its high incidence. Uncontrolled division and proliferation is one of the characteristics of tumor cells, which will promote rapid tumor growth and metastasis. Early mitotic inhibitor 1 (Emi1), ubiquitin-conjugating enzyme 10 (UBCH10) and CyclinB1 are important proteins involved in the regulation of cell cycle. In this study, the expression of Emi1, UBCH10 and CyclinB1 in ESCC tissues and adjacent normal tissues will be analyzed by immunohistochemistry and in-situ hybridization techniques, and their relationship with tumor proliferation and apoptosis will be analyzed. The results showed that Emi1, UBCH10 and CyclinB1 genes and proteins were highly expressed in tumor tissues, which were correlated with tumor grade, lymph node metastasis and pathological stage, and positively correlated with tumor proliferation. Emi1, UBCH10 and CyclinB1 are also positively correlated. It is speculated that Emi1, UBCH10 and CyclinB1 genes synergically promote tumor proliferation and inhibit apoptosis, which may be potential diagnostic and therapeutic targets for ESCC.
Introduction
Esophageal cancer is characterized by insidious onset, and patients often seek medical treatment in the middle and late stages, leading to poor prognosis and short survival time (1). Esophageal squamous cell carcinoma (ESCC) is a highly common subtype of esophageal carcinoma, so it is urgent to further explore the molecular pathology and the mechanism of malignant progression of ESCC. The uncontrolled proliferation of tumor cells is the result of dysregulation of cell cycle (2). Early mitotic inhibitor 1 (Emi1) mainly plays a role in promoting endogenous inhibitor of anaphase-promoting complex/cyclosome (APC/C), promoting the accumulation of S and G2 related mitotic Cyclin (3). Ubiquitin-conjugating enzyme 10 (UBCH10) is involved in the degradation of target proteins by the ubiquitin-proteasome system and thus plays a role in regulating the cell cycle (4). CyclinB1 belongs to the Cyclin family, which plays an important role in regulating the whole process of cell division and proliferation. Abnormal expression of cyclinB1 will cause the cell cycle to stall or exit, and the cell division and proliferation cannot be completed (5).
Currently, it is known that CyclinB1 is the target protein of ubiquitination protein degradation system, in which Emi1 and UBCH10 are participants. Studies have confirmed that Emi1, UBCH10 and CyclinB1 are abnormally expressed in a variety of tumors, and are closely related to the occurrence of tumors (6, 7). However, the expressions of Emi1, UBCH10 and CyclinB1 in ESCC, as well as whether there is interaction among them to jointly regulate the cell cycle process, are still unknown. Therefore, by exploring the expression of three proteins in ESCC and their correlation with tumor growth, we deeply understand the cell cycle process and regulatory mechanism of ESCC, hoping to provide a new theoretical basis for improving the therapeutic effect and prognosis of ESCC.
Methods
Tissue sample
All histological specimens were collected from confirmed ESCC and paracancer normal mucosal tissues from the Department of Pathology, the First Affiliated Hospital of Zhengzhou University from June 2020 to June 2021. All patients had not received radiotherapy or chemotherapy before surgery, and clinicopathologic data were complete. Among the 50 patients with ESCC, 29 were males and 21 were females. They ranged in age from 48 to 83, with a mean age of 60; 16 cases were classified as grade I-II and 34 cases as grade III. There were 23 cases with lymph node metastasis and 27 cases without lymph node metastasis. There were 27 cases of stage I to stage II and 23 cases of stage III to stage IV. Pathological staging was determined according to the TNM staging criteria for esophageal cancer (8th Edition) jointly published by the International Union Against Cancer (UICC) and the American Cancer Federation (AJCC) in 2017.
Reagents
Rabbit anti-human Emi1 polyclonal antibody and rabbit anti-human UBCH10 polyclonal antibody were purchased from Proteintech, United States. Rabbit anti-human CyclinB1 monoclonal antibody was purchased from Shanghai Biyuntian Biotechnology Co., LTD. Rabbit anti-human Ki-67 monoclonal antibody was purchased from Shanghai Gene Technology Co., LTD. TUNEL test kit was purchased from Jiangsu Kaiji Biotechnology Co., LTD. In situ hybridization biotin labeled probe was designed and synthesized by Shanghai GenePharma Technology Co., LTD.
Immunohistochemistry (IHC)
The slices were baked in a 61°C drying oven for 150min, and then dewaxed and hydrated. High pressure heating was used for antigen repair. Slices were added with appropriate 3% H2O2 drops, and then normal goat serum working solution was added. Add appropriate amount of primary antibody working solution and put the wet box in the refrigerator at 4°C overnight. On the second day, biotin labeled secondary antibody was added, then horseradish peroxidase labeled chain enzyme ovalbumin was added, and DAB working solution was added finally. ESCC tissue slices with known positive expressions of Emi1, UBCH10, CyclinB1 and Ki-67 were used as positive controls. PBS buffer will be used instead of primary antibody as the negative control.
Score by number of positive cells/percentage of observed cells. <1% is 0 point, 1%–25% is 1 point, 26%–50% is 2 point, 51%–75% is 3 point, >76% is 4 point. Score according to cell staining intensity: no color development is 0 point, light yellow is 1 point, brown and yellow is 2 point, tan is 3 point. Finally, the percentage score of positive cells was multiplied by the score of cell staining intensity to obtain the final score. The score ≤4 is negative, and the score >4 is positive.
In-situ hybridization (ISH)
The in-situ hybridization biotin labeled probe sequence is shown below. Emi1 probe sequence: 5′-CAACTATCCGAGGGTCGAGG-3′. UBCH10 probe sequence: 5′-CAGGGCTCCTGGCTGGTGACCTGCTT-3′. CyclinB1 probe sequence: 5′-CAGTGACTTCCCGACCCAGTAGGTATTT-3′.
The slices were baked in a 61°C drying oven for 150 min, and then dewaxed and hydrated. The slices were then dripped with an appropriate amount of 3% H2O2, followed by 0.3% Triton-X100. After pepsin was added, the pre-hybridization solution was added and incubated at 40°C for 3 h in a wet box. Biotin-labeled oligonucleotide probes were added and incubated overnight in a wet box at 42°C. On the second day, wash with SSC solution, add appropriate amount of sealing solution, and incubate in a wet box at 37°C for 30 min. Appropriate amount of SA-AP was added and incubated in a wet box at 37°C for 30 min. Appropriate amount of BCIP/NBT color developing working solution was added and incubated at 37°C in a wet box for 30–60 min. Appropriate amount of nuclear solid red dye solution was added to restain the nucleus for 5–15 min. ESCC slices with known positive expressions of Emi1, UBCH10 and CyclinB1 were used as positive controls. Hybridization solution without probe was used as negative control.
Score by number of positive cells/percentage of observed cells. <10% is 1 point, 11%–30% is 2 point, 31%–70% is 3 point, >70% is 4 point. Score according to cell staining intensity: no color development is 0 point, light blue is 1 point, darker purple blue is 2 point, deep purple blue is 3 point. Finally, the percentage score of positive cells was multiplied by the score of cell staining intensity to obtain the final score. The score <1 is negative, and the score ≥1 is positive.
In-situ end labeling (TUNEL)
The slices were baked in a 61°C drying oven for 150 min, and then dewaxed and hydrated. Microwave heating was used to repair antigen. The slices was dripped with 3% H2O2, and then the sealer was dripped. Add TdT enzyme working solution and incubate at 37°C for 60 min in the dark. Streptavidin-HRP working solution was added and incubated at 37°C for 30 min in the dark. DAB working solution was added and color was developed for 3–7 min. The known positive tissue slices were treated with Dnase I as positive control. Reaction solution without TDT enzyme was used as negative control.
The degree of apoptosis was suggested by the apoptosis index (AI), which was calculated according to the number of positive cells/percentage of observed cells: AI = number of positive cells/200*100%.
Statistical analysis
SPSS21.0 (United States) software was used for statistical analysis. Measurement data were expressed as ‾X ± S, and t-test was used to compare the differences between the two groups. Comparison of count data were performed by χ2 test or Fisher’s exact probability method, and correlation analysis was performed by Spearman method. Test level α = 0.05, p < 0.05 was considered statistically significant.
Results
The expression difference of Emi1, UBCH10 and CyclinB1 proteins in ESCC and paracancer tissues
The expressions of Emi1, UBCH10 and CyclinB1 proteins in ESCC and paracancer tissues were detected by IHC. The results showed that Emi1, UBCH10 and CyclinB1 proteins were highly expressed in ESCC tissues, as shown in Table 1 and Figure 1. There were significant differences in protein expression between ESCC and paracancer tissues (p < 0.05).
TABLE 1
| Group | Number of cases | Emi1 | UBCH10 | CyclinB1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| + | − | χ2 | p | + | − | χ2 | p | + | − | χ2 | p | ||
| ESCC | 50 | 43 | 7 | 46.314 | 0.000* | 44 | 6 | 54.782 | 0.000* | 43 | 7 | 54.782 | 0.000* |
| Paracancer | 50 | 9 | 41 | 7 | 43 | 6 | 44 | ||||||
The expression difference of Emi1, UBCH10 and CyclinB1 proteins in ESCC and paracancer tissues.
*p < 0.05.
FIGURE 1
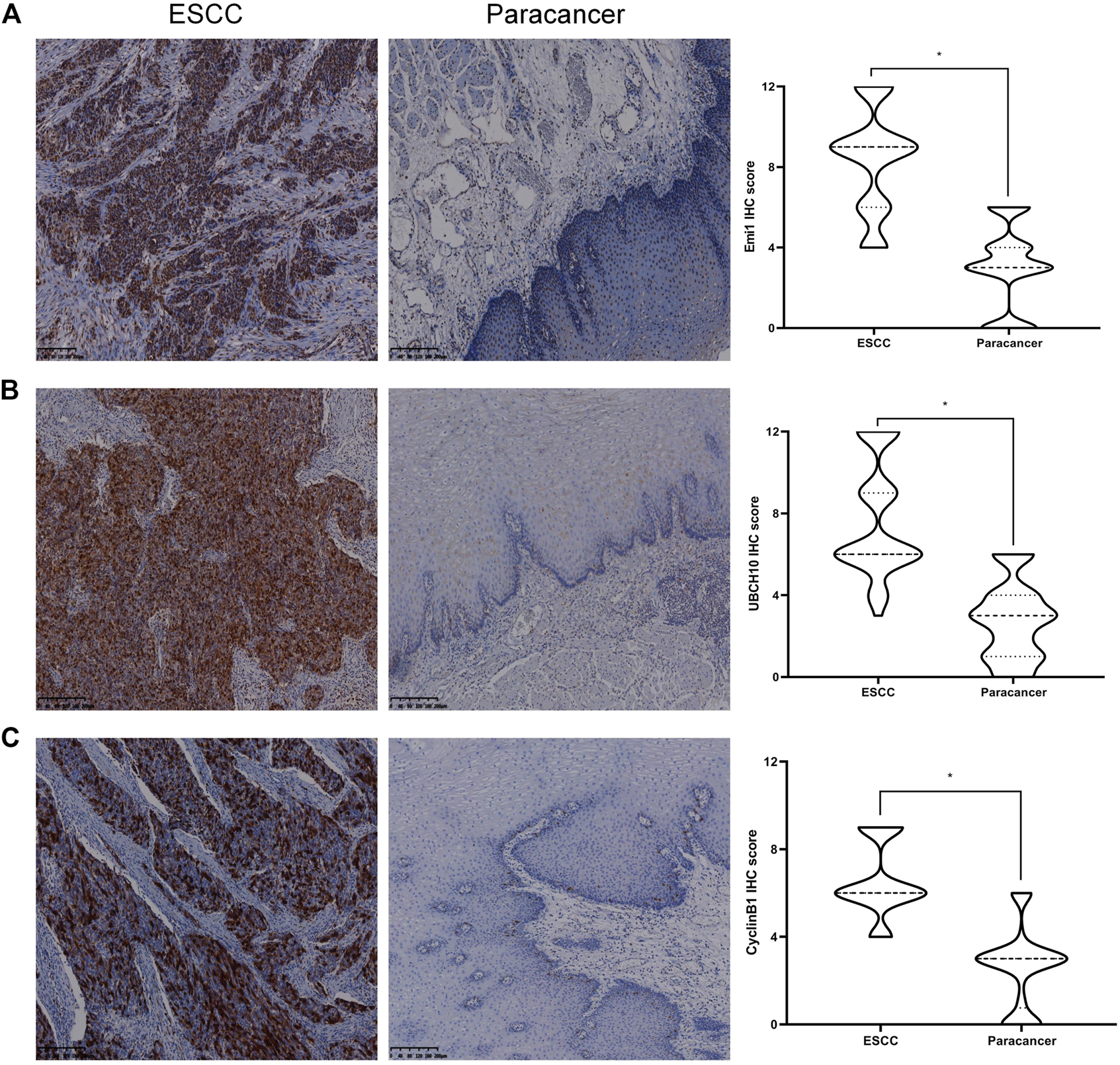
Expression of Emi1, UBCH10 and CyclinB1 proteins in ESCC and paracancer tissues. (A) Emi1 protein expression; (B) UBCH10 protein expression; (C) CyclinB1 protein expression (DAB color development, ×100).
The expression difference of Emi1, UBCH10 and CyclinB1 mRNA in ESCC and paracancer tissues
The expressions of Emi1, UBCH10 and CyclinB1 mRNA in ESCC and paracancer tissues were detected by ISH. The results showed that Emi1, UBCH10 and CyclinB1 mRNA were highly expressed in ESCC tissues, as shown in Table 2 and Figure 2. mRNA expression was significantly different between ESCC and paracancer tissues (p < 0.05).
TABLE 2
| Group | Number of cases | Emi1 | UBCH10 | CyclinB1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| + | − | χ2 | p | + | − | χ2 | p | + | − | χ2 | p | ||
| ESCC | 50 | 43 | 7 | 54.782 | 0.000* | 44 | 6 | 51.923 | 0.000* | 41 | 9 | 46.314 | 0.000* |
| Paracancer | 50 | 6 | 44 | 8 | 42 | 7 | 43 | ||||||
The expression difference of Emi1, UBCH10 and CyclinB1 mRNA in ESCC and paracancer tissues.
*p < 0.05.
FIGURE 2
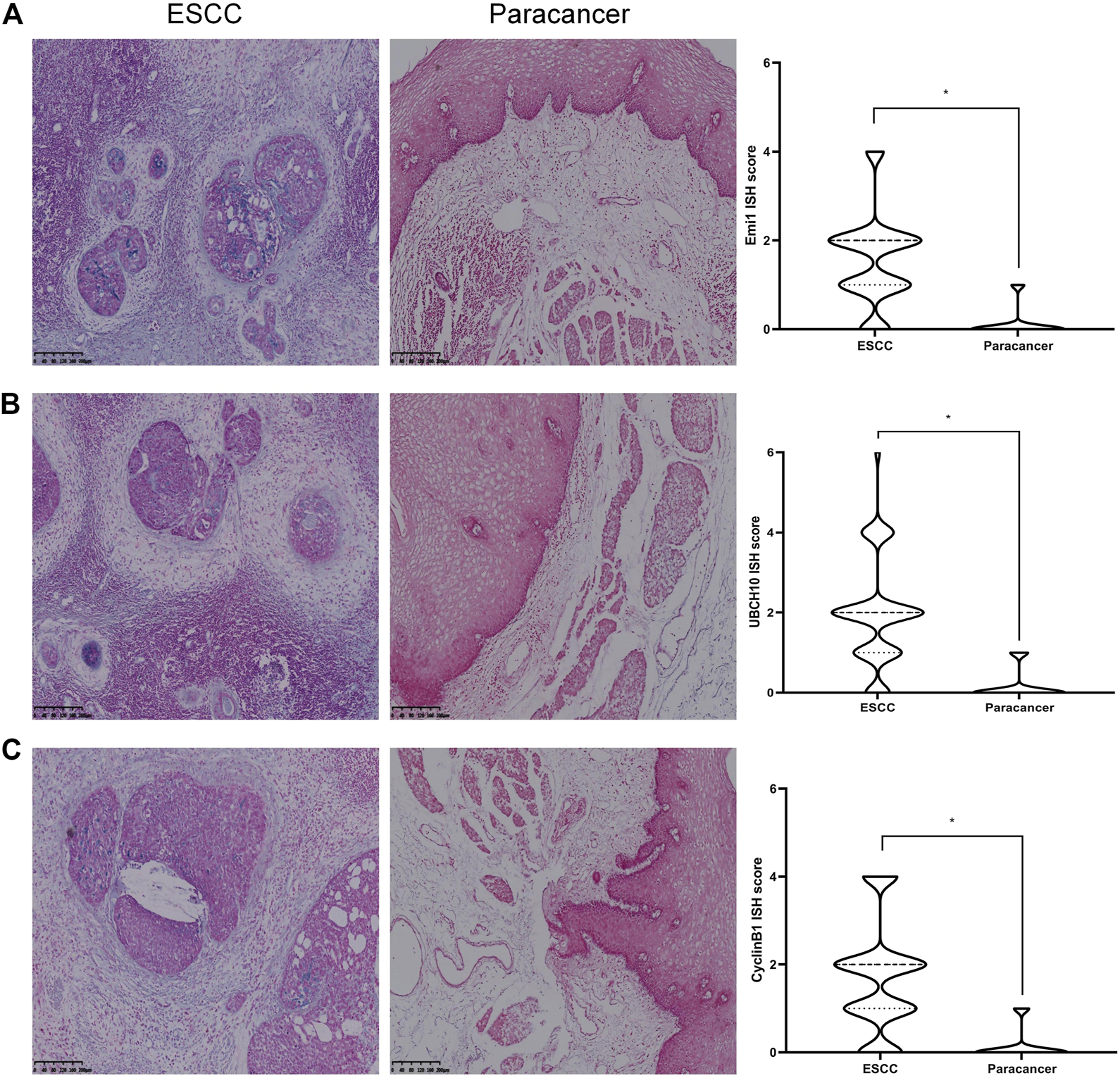
Expression of Emi1, UBCH10 and CyclinB1 mRNA in ESCC and paracancer tissues. (A) Emi1 mRNA expression; (B) UBCH10 mRNA expression; (C) CyclinB1 mRNA expression (BCIP/NBT color development, ×100).
Correlation between the expressions of Emi1, UBCH10, CyclinB1 and clinicopathological indexes
The correlation between Emi1, UBCH10, CyclinB1 and clinicopathological indexes was analyzed, including gender, age, tumor diameter, tissue grade, depth of invasion, lymph node metastasis, and pathological stage. The results showed that the expression of Emi1 protein and mRNA were correlated with tissue grade, lymph node metastasis and pathological stage (p < 0.05), as shown in Table 3. UBCH10 protein and mRNA expression were correlated with tissue grade, lymph node metastasis and pathological stage (p < 0.05), as shown in Table 4. CyclinB1 protein is correlated with tissue grade, lymph node metastasis, and pathological stage, while CyclinB1 mRNA is only correlated with tissue grade (p < 0.05). The difference between CyclinB1 protein and mRNA may be related to the small tissue sample size, as shown in Table 5.
TABLE 3
| Clinicopathological indexes | Number of cases | Emi1 protein | Emi1 mRNA | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Positive (%) | Negative (%) | χ2 | p | Positive (%) | Negative (%) | χ2 | p | ||
| Gender | |||||||||
| Male | 29 | 27 (93.10) | 2 (6.90) | 1.659 | 0.198 | 27 (93.10) | 2 (6.90) | 1.659 | 0.198 |
| Female | 21 | 16 (76.19) | 5 (23.81) | 16 (76.19) | 5 (23.81) | ||||
| Age | |||||||||
| <60 | 17 | 16 (94.12) | 1 (5.88) | 0.573 | 0.449 | 14 (82.35) | 3 (17.65) | 0.011 | 0.918 |
| ≥60 | 33 | 27 (81.82) | 6 (18.18) | 29 (87.88) | 4 (12.12) | ||||
| Tumor diameter (cm) | |||||||||
| <3 | 16 | 13 (81.25) | 3 (18.75) | 0.052 | 0.820 | 13 (81.25) | 3 (18.75) | 0.052 | 0.820 |
| ≥3 | 34 | 30 (88.24) | 4 (11.76) | 30 (88.24) | 4 (11.76) | ||||
| Tissue grade | |||||||||
| G1–G2 | 16 | 11 (68.75) | 5 (31.25) | 3.899 | 0.048* | 11 (68.75) | 5 (31.25) | 3.899 | 0.048* |
| G3 | 34 | 32 (94.12) | 2 (5.88) | 32 (94.12) | 2 (5.88) | ||||
| Depth of invasion | |||||||||
| T1–T2 | 21 | 17 (80.95) | 4 (19.05) | 0.214 | 0.644 | 18 (85.71) | 3 (14.29) | 0.000 | 1.000 |
| T3–T4 | 29 | 26 (89.66) | 3 (10.34) | 25 (86.21) | 4 (13.79) | ||||
| Lymph node metastasis | |||||||||
| N− | 27 | 20 (74.07) | 7 (25.93) | 6.934 | 0.011* | 20 (74.07) | 7 (25.93) | 6.934 | 0.011* |
| N+ | 23 | 23 (100) | 0 (0) | 23 (100) | 0 (0) | ||||
| Pathological stage | |||||||||
| I–II | 27 | 20 (74.07) | 7 (25.93) | 6.934 | 0.011* | 20 (74.07) | 7 (25.93) | 6.934 | 0.011* |
| III–IV | 23 | 23 (100) | 0 (0) | 23 (100) | 0 (0) | ||||
Correlation between the expressions of Emi1 and clinicopathological indexes.
N+, Lymph node metastasis; N−, No lymph node metastasis. *p < 0.05.
TABLE 4
| Clinicopathological indexes | Number of cases | UBCH10 protein | UBCH10 mRNA | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Positive (%) | Negative (%) | χ2 | p | Positive (%) | Negative (%) | χ2 | p | ||
| Gender | |||||||||
| Male | 29 | 28 (96.55) | 1 (3.45) | 3.048 | 0.081 | 26 (89.66) | 3 (10.34) | 0.000 | 1.000 |
| Female | 21 | 16 (76.19) | 5 (23.81) | 18 (85.71) | 3 (14.29) | ||||
| Age | |||||||||
| <60 | 17 | 15 (88.24) | 2 (11.76) | 0.001 | 1.000 | 15 (88.24) | 2 (11.76) | 0.000 | 1.000 |
| ≥60 | 33 | 29 (87.88) | 4 (12.12) | 29 (87.88) | 4 (12.12) | ||||
| Tumor diameter (cm) | |||||||||
| <3 | 16 | 15 (93.75) | 1 (6.25) | 0.154 | 0.695 | 13 (81.25) | 3 (18.75) | 0.293 | 0.588 |
| ≥3 | 34 | 29 (85.29) | 5 (14.71) | 31 (91.18) | 3 (8.82) | ||||
| Tissue grade | |||||||||
| G1–G2 | 16 | 11 (68.75) | 5 (31.25) | 5.794 | 0.016* | 11 (68.75) | 5 (31.25) | 5.794 | 0.016* |
| G3 | 34 | 33 (97.06) | 1 (2.94) | 33 (97.06) | 1 (2.94) | ||||
| Depth of invasion | |||||||||
| T1–T2 | 21 | 16 (76.19) | 5 (23.81) | 3.048 | 0.081 | 18 (85.71) | 3 (14.29) | 0.000 | 1.000 |
| T3–T4 | 29 | 28 (96.55) | 1 (3.45) | 26 (89.66) | 3 (10.34) | ||||
| Lymph node metastasis | |||||||||
| N− | 27 | 21 (77.78) | 6 (22.22) | 5.808 | 0.025* | 21 (77.78) | 6 (22.22) | 5.808 | 0.025* |
| N+ | 23 | 23 (100) | 0 (0) | 23 (100) | 0 (0) | ||||
| Pathological stage | |||||||||
| I–II | 27 | 21 (77.78) | 6 (22.22) | 5.808 | 0.025* | 21 (77.78) | 6 (22.22) | 5.808 | 0.025* |
| III–IV | 23 | 23 (100) | 0 (0) | 23 (100) | 0 (0) | ||||
Correlation between the expressions of UBCH10 and clinicopathological indexes.
N+, Lymph node metastasis; N−, No lymph node metastasis. *p < 0.05.
TABLE 5
| Clinicopathological indexes | Number of cases | CyclinB1 protein | CyclinB1 mRNA | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Positive (%) | Negative (%) | χ2 | p | Positive (%) | Negative (%) | χ2 | p | ||
| Gender | |||||||||
| Male | 29 | 27 (93.10) | 2 (6.90) | 1.659 | 0.198 | 26 (89.66) | 3 (10.34) | 1.646 | 0.200 |
| Female | 21 | 16 (76.19) | 5 (23.81) | 15 (71.43) | 6 (28.57) | ||||
| Age | |||||||||
| <60 | 17 | 16 (94.12) | 1 (5.88) | 0.573 | 0.449 | 14 (82.35) | 3 (17.65) | 0.000 | 1.000 |
| ≥60 | 33 | 27 (81.82) | 6 (18.18) | 27 (81.82) | 6 (18.18) | ||||
| Tumor diameter (cm) | |||||||||
| <3 | 16 | 14 (87.50) | 2 (12.50) | 0.000 | 1.000 | 12 (75.00) | 4 (25.00) | 0.239 | 0.625 |
| ≥3 | 34 | 29 (85.29) | 5 (14.71) | 29 (85.29) | 5 (14.71) | ||||
| Tissue grade | |||||||||
| G1–G2 | 16 | 11 (68.75) | 5 (31.25) | 3.899 | 0.048* | 9 (56.25) | 7 (43.75) | 8.160 | 0.004* |
| G3 | 34 | 32 (94.12) | 2 (5.88) | 32 (94.12) | 2 (5.88) | ||||
| Depth of invasion | |||||||||
| T1–T2 | 21 | 16 (76.19) | 5 (23.81) | 1.659 | 0.198 | 17 (80.95) | 4 (19.05) | 0.000 | 1.000 |
| T3–T4 | 29 | 27 (93.10) | 2 (6.90) | 24 (82.76) | 5 (17.24) | ||||
| Lymph node metastasis | |||||||||
| N− | 27 | 20 (74.07) | 7 (25.93) | 6.934 | 0.011* | 19 (70.37) | 8 (29.63) | 3.802 | 0.051 |
| N+ | 23 | 23 (100) | 0 (0) | 22 (95.65) | 1 (4.35) | ||||
| Pathological stage | |||||||||
| I–II | 27 | 20 (74.07) | 7 (25.93) | 6.934 | 0.011* | 19 (70.37) | 8 (29.63) | 3.802 | 0.051 |
| III–IV | 23 | 23 (100) | 0 (0) | 22 (95.65) | 1 (4.35) | ||||
Correlation between the expressions of CyclinB1 and clinicopathological indexes.
N+, Lymph node metastasis; N−, No lymph node metastasis. *p < 0.05.
Correlation of Emi1, UBCH10 and CyclinB1 expression
The correlation of protein expression among Emi1, UBCH10 and CyclinB1 in ESCC tissues was analyzed, as shown in Figure 3. The results showed that Emi1 was positively correlated with UBCH10 protein expression (r = 0.5418, p < 0.0001). Emi1 was positively correlated with the expression of CyclinB1 (r = 0.5539, p < 0.0001). UBCH10 was positively correlated with the expression of CyclinB1 (r = 0.6020, p < 0.0001).
FIGURE 3
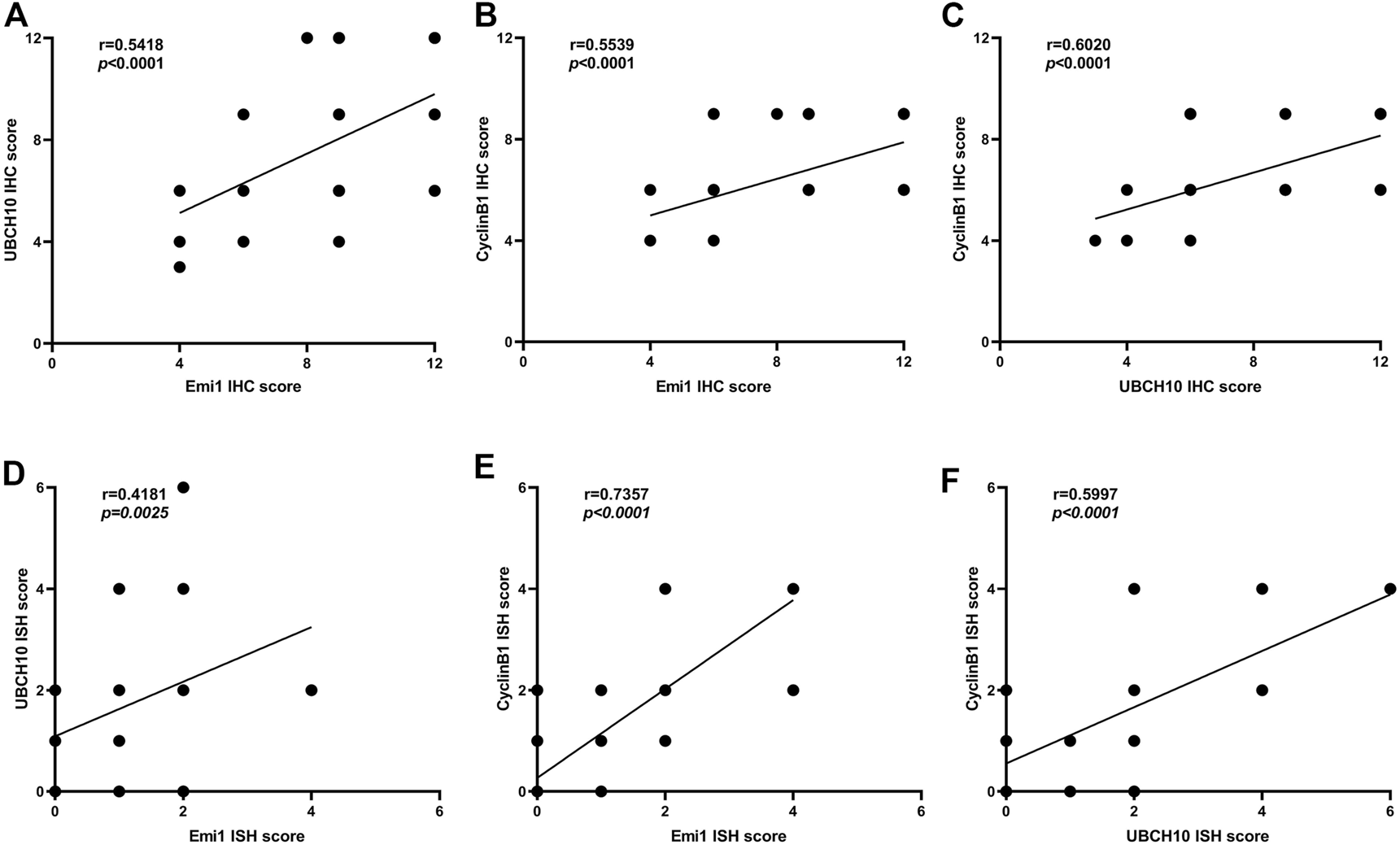
Correlation between Emi1, UBCH10 and CyclinB1 expression (A dot may represent multiple samples with the same score). (A) Protein expression between Emi1 and UBCH10; (B) Protein expression between Emi1 and CyclinB1; (C) Protein expression between UBCH10 and CyclinB1; (D) mRNA expression between Emi1 and UBCH10; (E) mRNA expression between Emi1 and CyclinB1; (F) mRNA expression between UBCH10 and CyclinB1.
The mRNA expression correlation among Emi1, UBCH10 and CyclinB1 in ESCC tissues was further analyzed, as shown in Figure 3. The results showed that Emi1 was positively correlated with UBCH10 mRNA expression (r = 0.4181, p = 0.0025). Emi1 was positively correlated with the expression of CyclinB1 mRNA (r = 0.7357, p < 0.0001). UBCH10 was positively correlated with the expression of CyclinB1 mRNA (r = 0.5997, p < 0.0001).
Proliferation and apoptosis in ESCC and paracancer tissues
The expression of Ki-67 protein in ESCC and paracancer tissues was detected by IHC. Ki-67 is a nuclear antigen closely related to cell proliferation. The proliferation index was determined by evaluating the expression of Ki-67 in ESCC and paracancer tissues, as shown in Table 6 and Figure 4. The results showed that the ESCC tissue presented a very obvious high proliferation index, with a mean of 60.40%, while the proliferation index of the paracancer tissue was significantly reduced, about 11.90%, compared with the tumor tissue (p < 0.05).
TABLE 6
| Group | Number of cases | Proliferation index | Apoptosis index | ||
|---|---|---|---|---|---|
| Mean ± s.d | p | Mean ± s.d | p | ||
| ESCC | 50 | 60.40 ± 15.08 | 0.000* | 29.60 ± 20.89 | 0.002* |
| Paracancer | 50 | 11.90 ± 6.84 | 42.20 ± 18.98 | ||
Proliferation index and apoptosis index in ESCC and paracancer tissue (‾X ± S) (%).
*p < 0.05.
FIGURE 4
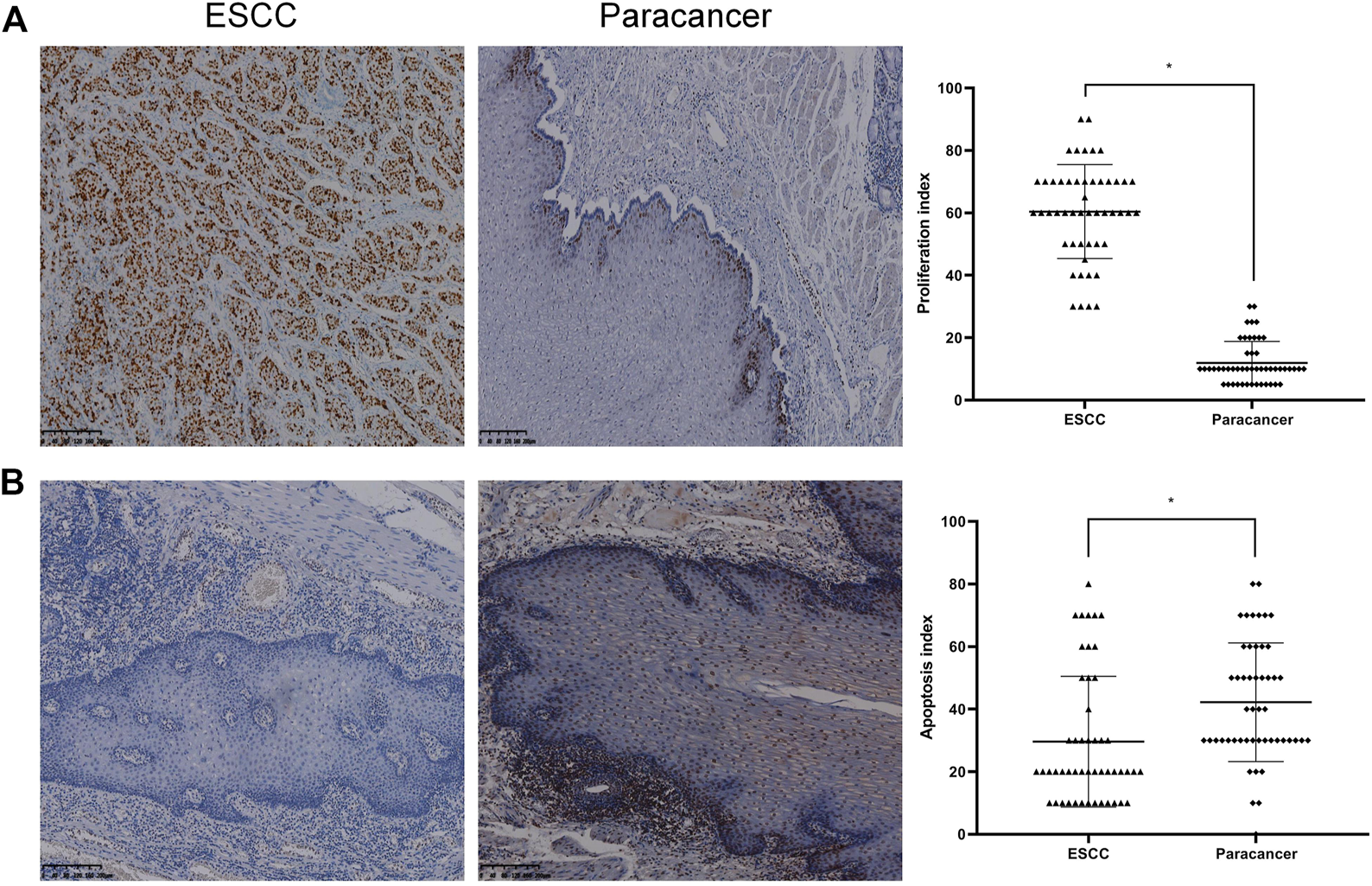
Proliferation and apoptosis in ESCC and paracancer tissues [(A) Proliferation in ESCC and paracancer tissue; (B) Apoptosis in ESCC and paracancer tissues] (DAB color development, ×100).
Apoptosis in ESCC and paracancer tissues was determined by TUNEL technique, as shown in Table 6 and Figure 4. The results showed that the apoptotic index was about 29.60% in ESCC tissue and 42.20% in paracarcinoma tissue, and the apoptotic index was lower in ESCC tissue. The proliferation and apoptosis indexes of ESCC and paracancer tissues were significantly different (p < 0.05).
Correlation between Emi1, UBCH10, CyclinB1 expression and tumor proliferation
The correlation between Emi1, UBCH10 and CyclinB1 protein expression and proliferation index in ESCC tissues was analyzed, as shown in Figure 5. The results showed that the protein expression of Emi1, UBCH10 and CyclinB1 was positively correlated with the proliferation index (r = 0.4561, p = 0.0009) (r = 0.4082, p = 0.0033) (r = 0.4300, p = 0.0018).
FIGURE 5
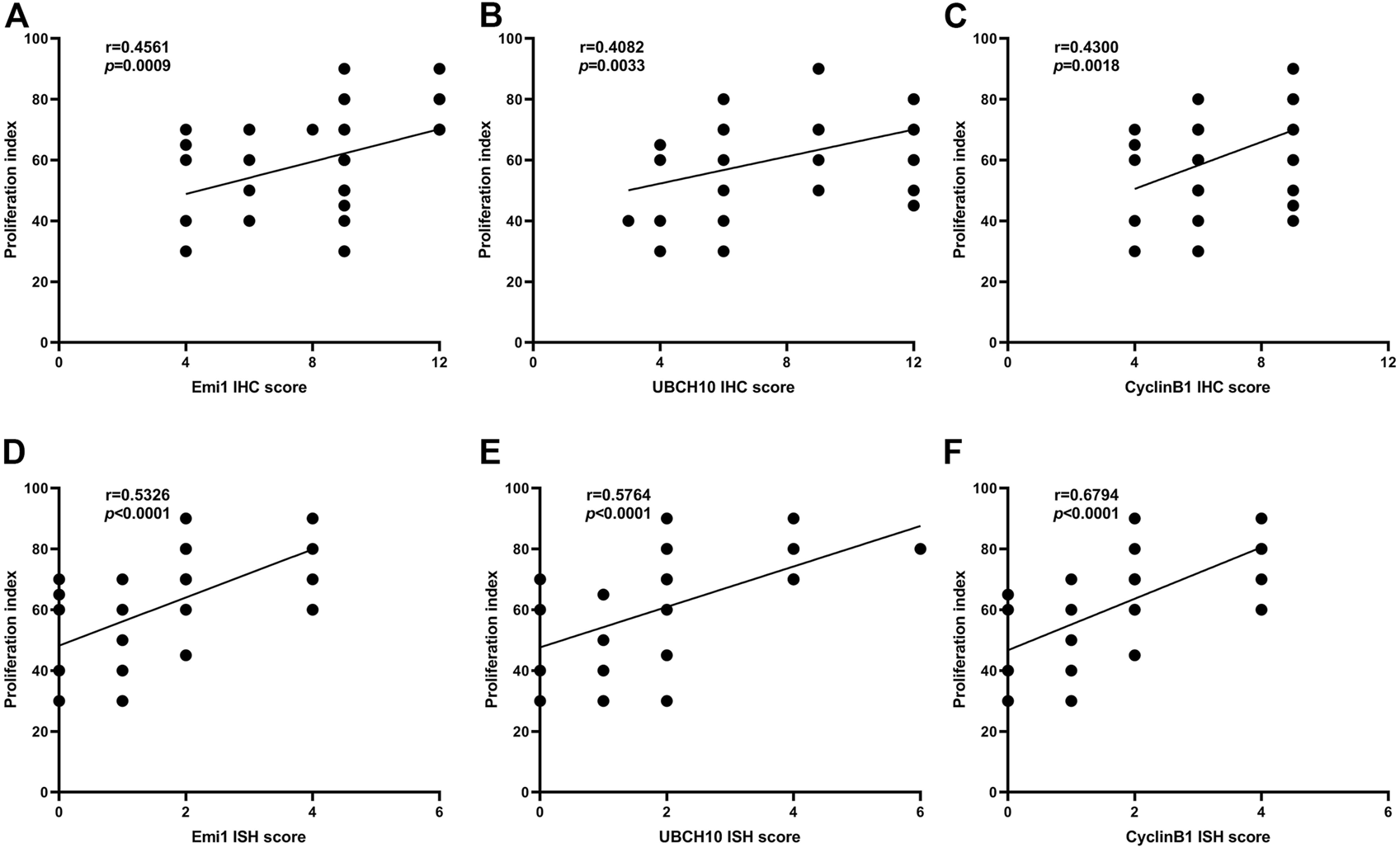
Correlation between Emi1, UBCH10, CyclinB1 and proliferation index (A dot may represent multiple samples with the same score). (A) Correlation between Emi1 protein and proliferation index; (B) Correlation between UBCH10 protein and proliferation index; (C) Correlation between CyclinB1 protein and proliferation index; (D) Correlation between Emi1 mRNA and proliferation index; (E) Correlation between UBCH10 mRNA and proliferation index; (F) Correlation between CyclinB1 mRNA and proliferation index.
The correlation between Emi1, UBCH10 and CyclinB1 mRNA expression and proliferation index in ESCC tissues was analyzed, as shown in Figure 5. The results showed that the mRNA expression of Emi1, UBCH10 and CyclinB1 was positively correlated with the proliferation index (r = 0.5326, p < 0.0001) (r = 0.5764, p < 0.0001) (r = 0.6794, p < 0.0001).
Correlation between expression of Emi1, UBCH10, CyclinB1 and tumor apoptosis
The correlation between Emi1, UBCH10, CyclinB1 protein expression and apoptosis index in ESCC tissues was analyzed, as shown in Figure 6. The results showed that the protein expressions of Emi1, UBCH10 and CyclinB1 were negatively correlated with the apoptosis index (r = −0.5737, p < 0.0001) (r = −0.4178, p = 0.0025) (r = −0.4939, p = 0.0018).
FIGURE 6
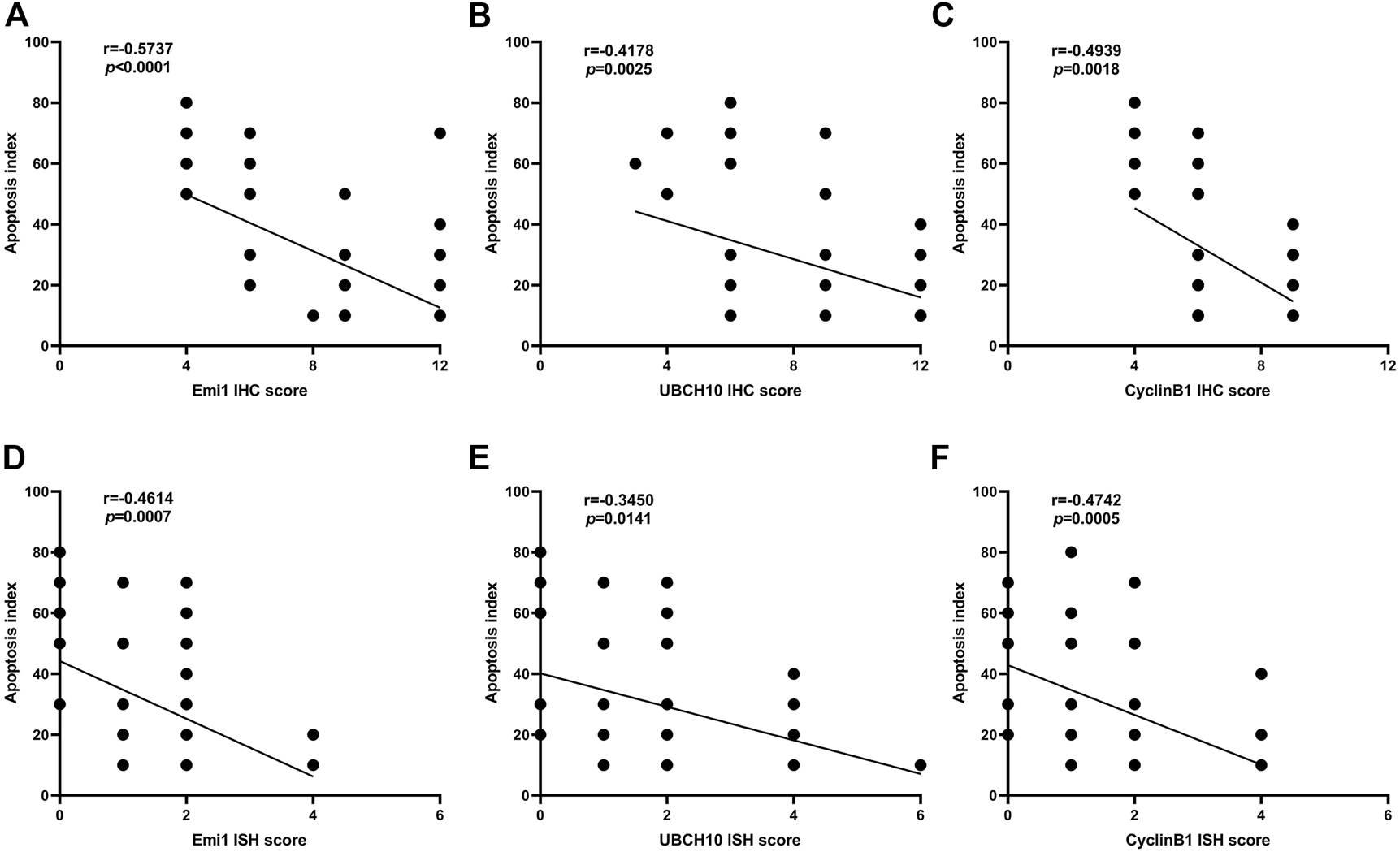
Correlation between Emi1, UBCH10, CyclinB1 and apoptosis index (A dot may represent multiple samples with the same score). (A) Correlation between Emi1 protein and apoptosis index; (B) Correlation between UBCH10 protein and apoptosis index; (C) Correlation between CyclinB1 protein and apoptosis index; (D) Correlation between Emi1 mRNA and apoptosis index; (E) Correlation between UBCH10 mRNA and apoptosis index; (F) Correlation between CyclinB1 mRNA and apoptosis index.
The correlation between Emi1, UBCH10 and CyclinB1 mRNA expression and apoptosis index in ESCC tissues was analyzed, as shown in Figure 6. The results showed that the mRNA expressions of Emi1, UBCH10 and CyclinB1 were negatively correlated with the apoptosis index (r = −0.4614, p = 0.0007) (r = −0.3450, p = 0.0141) (r = −0.4742, p = 0.0005).
Discussion
Although a series of early screening work has been carried out in areas with high incidence of esophageal cancer, showing preliminary tumor prevention and control effect, the tumor burden of esophageal cancer is still very large. Finding new genes and proteins that may play an important role in the occurrence and development of ESCC and elucidating their related molecular mechanisms will be of great significance in the diagnosis and treatment of ESCC. Normal cells proliferate through mitosis and are regulated by a variety of cyclins, Cyclin-dependent protein kinases (CDKs), cell cycle checkpoints and cell cycle signaling pathways (8). The level of cyclin determines the mitotic process of cells. When the level of cyclin decreases, the cell cycle will actively stop or even exit the mitotic process. Protein ubiquitination plays an important role in the stabilization of cyclin levels (9).
Emi1 was initially thought to be a protein involved in the regulation of cell cycle. However, with the development of tumor research, researchers found that the “identity” of Emi1 in the development of tumor was actually oncogene (10). However, there are few reports on the expression of Emi1 in ESCC tissues, so this study started from exploring the expression of Emi1 gene in ESCC. We found that Emi1 mRNA and protein expressions in ESCC tumor tissues were higher than those in paracancer normal esophageal mucosa tissues, confirming the view that Emi1 is an oncogenic gene. Moreover, Emi1 expression was correlated with tumor differentiation, lymph node metastasis, and pathological stage, and was positively correlated with proliferation and negatively correlated with apoptosis, suggesting that Emi1 played an important role in the malignant process of ESCC. The findings of Emi1 in breast cancer are similar to our findings, Emi1 mediates the anti-apoptotic and pro-proliferative carcinogenicity of Skp2 through PI3K/Akt signaling pathway (11).
UBCH10 is an active protein in ubiquitin-proteasome. However, in addition to regulating ubiquitination degradation of protein, UBCH10 is also closely related to tumor proliferation and metastasis, and may be a new tumor marker or therapeutic target (4). Our study found that UBCH10 was significantly highly expressed in ESCC, which was related to tumor differentiation, lymph node metastasis, and pathological stage, and was positively correlated with tumor proliferation index and negatively correlated with apoptosis index, suggesting that UBCH10 could promote tumor proliferation and inhibit tumor apoptosis in ESCC. Compared with ESCC, UBCH10 has been extensively studied in other tumors. In glioma, UBCH10 expression increases with the increase of tumor malignancy. After siRNA silencing UBCH10, glioma cells show growth inhibition, cell cycle arrest and increased apoptosis (12, 13). UBCH10 in lung cancer is also related to tumor differentiation and patient survival, and UBCH10 can lead to P53 and EGFR gene mutations, resulting in loss of tumor inhibitory effect of P53 gene and enhancement of growth promoting effect of EGFR, while the proliferation of lung cancer cells and drug resistance of chemotherapy drugs are weakened after UBCH10 gene silencing (14, 15). UBCH10 is associated with ER and Ki-67 in breast cancer. Silencing UBCH10 can inhibit the proliferation of tumor cells and increase the sensitivity to chemotherapy. UBCH10 has also been detected in circulating tumor cells, suggesting that it can be used as an indicator for early screening and diagnosis of breast cancer (16).
CyclinB1 is a “star molecule” in the cyclin family and a core protein that regulates the G2 phase of the cell cycle. Similar to UBCH10, CyclinB1 not only has cell cycle regulation function, but also is an oncogenic gene associated with abnormal tumor proliferation. Through the detection of CyclinB1 mRNA and protein in ESCC tissues, it was found that the high expression of CyclinB1 in ESCC tissues was positively correlated with the proliferation index and negatively correlated with the apoptosis index. The expression of CyclinB1 protein was correlated with tumor grade, lymph node metastasis and pathological stage. Other researchers have found evidence that confirms our results, The expression of CyclinB1 was downregulated in cervical cancer cells, and it was found that tumor progression was inhibited and cell cycle was stagnated in G2/M phase, which ultimately delayed the process of tumor development (17, 18). When the level of CyclinB1 was reduced in breast cancer cells, the proliferation and migration of tumor cells were inhibited, and the cell cycle was stagnated in G2/M phase (7).
Emi1 is involved in cell cycle regulation by acting as an endogenous inhibitor of APC/C and hindering the degradation of its substrates by APC/C (19). UBCH10, as a member of ubiquitin binding enzyme E2 family, binds to ubiquitin ligase E3 to form a complex to initiate ubiquitin-proteasome degradation pathway, degrades APC/C substrate CyclinB, and finally plays a role in regulating cell cycle (20). CyclinB1 is mainly involved in the regulation of G2 phase of the cell cycle. In late mitosis, CyclinB1 combines with CDK1 to form a complex, Cyclinb1-CDK1, which phosphorylates APC/C and then promotes the degradation of CyclinB1 (21). Emi1, UBCH10 and CyclinB1 are all participants in the cell cycle regulation mechanism and are key points in the APC/C molecular mechanism network. By analyzing the correlation between the expressions of Emi1, UBCH10 and CyclinB1, the results confirmed that there was a positive correlation between Emi1, UBCH10 and CyclinB1 at both protein level and mRNA level. The degradation of cyclins is mainly dependent on ubiquitination, among which APC/C is an important ubiquitin-binding enzyme E3, and the degradation substrates of APC/C include a variety of proteins such as CyclinA and CyclinB. The silencing of Emi1 in cells will cause a large number of ubiquitination substrate proteins of APC/C, resulting in ubiquitination degradation of CyclinA and CyclinB, and insufficient accumulation of cyclin in cells, which cannot enter the next phase, and eventually lead to cell cycle arrest (22). However, overexpression of Emi1 in cells will block the catalytic site of APC/C and competitively prevent APC/C substrate proteins from binding to APC/C co-receptors, resulting in increased CyclinA and CyclinB levels and disorder of cell proliferation cycle (19). Normal cell cycle arrest and cell physiological apoptosis can not be carried out, which further leads to malignant cell proliferation, promotes cell proliferation and inhibits cell apoptosis. In-depth analysis of the inhibitory effect and mechanism of Emi1 revealed that Emi1 protein contains a variety of domains that block the substrate binding site of APC/C and inhibit the formation of ubiquitin chains (23). As ubiquitin binding enzyme E2, UBCH10 has the function of forming and extending ubiquitin chains, which can connect the substrate protein and ubiquitin ligase E3 through the ubiquitin chain, label ubiquitin on the substrate protein, and initiate the ubiquitination degradation process (24). In normal cells, Emi1 can effectively inhibit UBCH10 ubiquitin chain extension, thus effectively stabilizing the level of substrate protein (25).
At present, the specific molecular mechanism of Emi1, UBCH10 and CyclinB1 genes in promoting tumor proliferation and inhibiting apoptosis has not been reported. Combined with our experimental results, we conjectured that Emi1 reduced UBCH10 consumption by inhibiting ubiquitin chain extension between UBCH10 and CyclinB1, and inhibited the ubiquitination degradation of CyclinB1 protein, resulting in high expression of Emi1, UBCH10 and CyclinB1 in tumor tissues. Cell cycle regulation is a complex and huge mechanism network. In addition to the influence of UBCH10 and CyclinB1 proteins, Emi1 may also have other molecular effects, which requires further discussion.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving human participants were reviewed and approved by Life Science Ethics Review Committee of Zhengzhou University. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
KC and MS designed the research study and approved the final version of the manuscript for publication. HL completed the experiment. CY conducted the research and wrote the paper. All authors read and approved the final manuscript.
Funding
National Natural Science Foundation of China (81873455); Training Program for young and middle-aged Health Science and Technology innovation Leaders in Henan Province (YXKC2022015).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.por-journal.com/articles/10.3389/pore.2023.1611081/full#supplementary-material
References
1.
Arnold M Abnet CC Neale RE Vignat J Giovannucci EL McGlynn KA et al Global burden of 5 major types of gastrointestinal cancer. Gastroenterology (2020) 159:335–49. 10.1053/j.gastro.2020.02.068
2.
Lezaja A Altmeyer M . Dealing with DNA lesions: When one cell cycle is not enough. Curr Opin Cel Biol (2021) 70:27–36. 10.1016/j.ceb.2020.11.001
3.
Zhao Y Tang Q Ni R Huang X Wang Y Lu C et al Early mitotic inhibitor-1, an anaphase-promoting complex/cyclosome inhibitor, can control tumor cell proliferation in hepatocellular carcinoma: Correlation with Skp2 stability and degradation of p27(kip1). Hum Pathol (2013) 44:365–73. 10.1016/j.humpath.2012.03.030
4.
Presta I Novellino F Donato A La Torre D Palleria C Russo E et al UbcH10 a major actor in cancerogenesis and a potential tool for diagnosis and therapy. Int J Mol Sci (2020) 21:2041. 10.3390/ijms21062041
5.
Wood DJ Endicott JA . Structural insights into the functional diversity of the CDK-cyclin family. Open Biol (2018) 8:180112. 10.1098/rsob.180112
6.
Lei H Wang K Jiang T Lu J Dong X Wang F et al KIAA0101 and UbcH10 interact to regulate non-small cell lung cancer cell proliferation by disrupting the function of the spindle assembly checkpoint. BMC Cancer (2020) 20:957. 10.1186/s12885-020-07463-3
7.
Liu B Liu Y Wang Y Xie C Gan M Han T et al CyclinB1 deubiquitination by USP14 regulates cell cycle progression in breast cancer. Pathol Res Pract (2019) 215:152592. 10.1016/j.prp.2019.152592
8.
Sun Y Liu Y Ma X Hu H . The influence of cell cycle regulation on chemotherapy. Int J Mol Sci (2021) 22:6923. 10.3390/ijms22136923
9.
Peters JM . The anaphase promoting complex/cyclosome: A machine designed to destroy. Nat Rev Mol Cel Biol. (2006) 7:644–56. 10.1038/nrm1988
10.
Yamano H . EMI1, a three-in-one ubiquitylation inhibitor. Nat Struct Mol Biol (2013) 20:773–4. 10.1038/nsmb.2626
11.
Liu X Wang H Ma J Xu J Sheng C Yang S et al The expression and prognosis of Emi1 and Skp2 in breast carcinoma: Associated with PI3K/akt pathway and cell proliferation. Med Oncol (2013) 30:735. 10.1007/s12032-013-0735-0
12.
Donato G Iofrida G Lavano A Volpentesta G Signorelli F Pallante PL et al Analysis of UbcH10 expression represents a useful tool for the diagnosis and therapy of astrocytic tumors. Clin Neuropathol (2008) 27:219–23. 10.5414/npp27219
13.
Jiang L Bao Y Luo C Hu G Huang C Ding X et al Knockdown of ubiquitin-conjugating enzyme E2C/UbcH10 expression by RNA interference inhibits glioma cell proliferation and enhances cell apoptosis in vitro. J Cancer Res Clin Oncol (2010) 136:211–7. 10.1007/s00432-009-0651-z
14.
Perrotta I Bruno L Maltese L Russo E Donato A Donato G . Immunohistochemical analysis of the ubiquitin-conjugating enzyme UbcH10 in lung cancer: A useful tool for diagnosis and therapy. J Histochem Cytochem (2012) 60:359–65. 10.1369/0022155412439717
15.
Pallante P Malapelle U Berlingieri MT Bellevicine C Sepe R Federico A et al UbcH10 overexpression in human lung carcinomas and its correlation with EGFR and p53 mutational status. Eur J Cancer (2013) 49:1117–26. 10.1016/j.ejca.2012.09.033
16.
Wang C Pan YH Shan M Xu M Bao JL Zhao LM . Knockdown of UbcH10 enhances the chemosensitivity of dual drug resistant breast cancer cells to epirubicin and docetaxel. Int J Mol Sci (2015) 16:4698–712. 10.3390/ijms16034698
17.
Zhu B Zhang Q Wu Y Luo J Zheng X Xu L et al SNAP23 suppresses cervical cancer progression via modulating the cell cycle. Gene (2018) 673:217–24. 10.1016/j.gene.2018.06.028
18.
Cheng YM Tsai CC Hsu YC . Sulforaphane, a dietary isothiocyanate, induces G₂/M arrest in cervical cancer cells through CyclinB1 downregulation and gadd45β/CDC2 association. Int J Mol Sci (2016) 17:1530. 10.3390/ijms17091530
19.
Vaidyanathan S Cato K Tang L Pavey S Haass NK Gabrielli BG et al In vivo overexpression of Emi1 promotes chromosome instability and tumorigenesis. Oncogene (2016) 35:5446–55. 10.1038/onc.2016.94
20.
van Ree JH Jeganathan KB Malureanu L van Deursen JM . Overexpression of the E2 ubiquitin-conjugating enzyme UbcH10 causes chromosome missegregation and tumor formation. J Cel Biol (2010) 188:83–100. 10.1083/jcb.200906147
21.
Gavet O Pines J . Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev Cel (2010) 18:533–43. 10.1016/j.devcel.2010.02.013
22.
Lee H Lee DJ Oh SP Park HD Nam HH Kim JM et al Mouse emi1 has an essential function in mitotic progression during early embryogenesis. Mol Cel Biol (2006) 26:5373–81. 10.1128/mcb.00043-06
23.
Frye JJ Brown NG Petzold G Watson ER Grace CR Nourse A et al Electron microscopy structure of human APC/C(CDH1)-EMI1 reveals multimodal mechanism of E3 ligase shutdown. Nat Struct Mol Biol (2013) 20:827–35. 10.1038/nsmb.2593
24.
Bonacci T Emanuele MJ . Impressionist portraits of mitotic exit: APC/C, K11-linked ubiquitin chains and cezanne. Cell Cycle (Georgetown, Tex.) (2019) 18:652–60. 10.1080/15384101.2019.1593646
25.
Wang W Kirschner MW . Emi1 preferentially inhibits ubiquitin chain elongation by the anaphase-promoting complex. Nat Cel Biol (2013) 15:797–806. 10.1038/ncb2755
Summary
Keywords
tumor proliferation, Emi1, UBCH10, CyclinB1, tumor apoptosis
Citation
Li H, Yang C, Chen K and Sun M (2023) Expression significance of Emi1, UBCH10 and CyclinB1 in esophageal squamous cell carcinoma. Pathol. Oncol. Res. 29:1611081. doi: 10.3389/pore.2023.1611081
Received
27 January 2023
Accepted
11 April 2023
Published
24 April 2023
Volume
29 - 2023
Edited by
Anna Sebestyén, Semmelweis University, Hungary
Updates
Copyright
© 2023 Li, Yang, Chen and Sun.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kuisheng Chen, chenksh2002@163.com; Miaomiao Sun, sunmiaomiaohd@163.com
†These authors have contributed equally to this work and share first authorship
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.