Abstract
KRAS and BRAF mutations are currently thought to be mutually exclusive as their co-occurrence is extremely rare. Therefore, clinicopathological and molecular characteristics of colorectal carcinoma with KRAS/BRAF double mutations are unclear. We aimed to investigate the frequency and clinicopathological characteristics of double-mutant colorectal carcinoma and its differences from KRAS/BRAF single-mutant colorectal carcinoma using bioinformatics tools. We estimated the KRAS/BRAF double mutation frequency in the whole exon and coding sequences via bioinformatic analyses of three datasets from cBioPortal. We compared the clinicopathological characteristics, microsatellite instability status, BRAF classification, and tumor mutation burden of patients harboring the double mutants with those of patients harboring KRAS or BRAF single mutations. We integrated three large datasets and found that the frequency of the KRAS/BRAF double mutation in the dataset was 1.2% (29/2347). The double mutation occurred more frequently in males, with a slightly higher occurrence in the right side of the colon. Sex, histological type, histological grade, microsatellite instability, and tumor mutation burden of the patients harboring KRAS-mutant, BRAF-mutant, and double-mutant colorectal carcinoma varied significantly. The frequency of double-mutant colorectal carcinoma was 60 times higher than that previously reported. Significantly fewer double-mutant colorectal carcinoma cases were classified as BRAF class 1 and more were classified as unknown. Our findings indicate that the biological characteristics of double-mutant tumors are different from those of single-mutant tumors.
Introduction
Colorectal cancer (CRC) is the second-most common cancer in women and the third-most common cancer in men [1]. CRC progresses through several steps associated with specific genetic and epigenetic alterations in various oncogenes and tumor suppressor genes [2]. Kirsten rat sarcoma viral oncogene homolog (KRAS) and v-RAF murine sarcoma viral oncogene homolog B1 (BRAF) are the major oncogenic drivers of CRC [3]. Approximately 30–45% of patients with CRC harbor KRAS mutations and 5–20% harbor BRAF mutations [4]. KRAS and BRAF encode proteins involved in the Ras–Raf–MEK–ERK signaling pathway. KRAS can also activate other signaling pathways, such as the PIK3CA–AKT–mTOR pathway, which regulates protein translation and cell survival [5] Therefore, gain-of-function KRAS and BRAF mutations activate these pathways that act as molecular switches leading to cellular growth and proliferation and are associated with primary resistance to epidermal growth factor receptor (EGFR) inhibitors [6,7]. Recent studies have shown that BRAF V600-mutated CRC and BRAF non-V600-mutated CRC have different prognoses and different sensitivities to drugs; furthermore, the proposed BRAF mutations can be grouped into three classes (1, 2, and 3) [8,9] Currently, combinatorial therapy with cytotoxic chemotherapeutic agents and molecular targeted drugs (bevacizumab) are recommended as the first-line therapy for KRAS/BRAF-mutant CRC [10].
G12D, G12V, and G13D, the most common missense KRAS mutations and BRAF V600E have been recognized as being mutually exclusive [11,12]. In previous studies, the double KRAS/BRAF mutation frequency was 0.02% (1/4,170) [13–19]. However, reports regarding the occurrence of KRAS and BRAF double mutants have recently emerged [20–25]. To the best of our knowledge, only 11 cases have presented the co-occurrence of KRAS and BRAF mutations, indicating that this mutation is extremely rare. Owing to the rarity of the KRAS/BRAF double mutation, the clinicopathological and molecular characteristics of KRAS/BRAF double-mutant tumors and differences in the biology of KRAS or BRAF single-mutant CRC and KRAS/BRAF double-mutant CRC remain unknown.
In this study, we analyzed the frequency of KRAS/BRAF double mutations and the methods used for detecting these double mutations and determined the frequency of double-mutant CRC from three public datasets using bioinformatic tools. Additionally, we examined the clinicopathological features, microsatellite instability (MSI) status, tumor mutation burden (TMB), CpG island methylator phenotype (CIMP), BRAF classification, and the clinicopathological and molecular differences between CRCs with single KRAS or BRAF mutations and those with double mutations. To our knowledge, this is the first study to determine the KRAS/BRAF double mutation frequency in a large dataset. This study demonstrated the frequency of double-mutant colorectal carcinoma and clarified the clinicopathological and molecular features of double-mutant CRC.
Materials and Methods
Data Collection
Genomic and clinical data associated with tumor samples from patients with colorectal adenocarcinoma (The Cancer Genome Atlas [TCGA] PanCancer Atlas; n = 594), metastatic CRC [Memorial Sloan-Kettering Cancer Center (MSKCC), n = 1,134] [26], and colorectal adenocarcinoma [Dana-Farber Cancer Institute (DFCI), n = 619] [27] were accessed online via the cBioPortal. We extracted datasets for KRAS mutation, BRAF mutation, and KRAS/BRAF double mutation from all the samples (n = 2347), including TCGA, MSKCC, and DFCI tumor samples combined. Clinicopathological features, including age, sex, tumor location, histological type, grade (G1, G2, and G3), tumor–node–metastasis classification (only TCGA), stage, CIMP (only DFCI) and overall survival data were obtained from TCGA and MSKCC via the cBioPortal. Additionally, a list of amino-acid changes and information regarding the pathological significance of each KRAS or BRAF mutation were accessed using COSMIC [28]. Allele frequency was assessed using cBioPortal (Supplementary Table S1).
Mutation Data
In TCGA, MutSig2CV was applied to quality-controlled mutation data to evaluate the significance of the mutated genes and estimate the mutation densities of samples. MutSig2CV [29] combines evidence from the background mutation rate, clustering of mutation on hotspots, and conservation of mutated sites to calculate false discovery rates (q-values). Genes with q-value <0.1 were considered significant [30].
In MSKCC, the thresholds on the coverage depth, number of mutant reads, and variant frequency for rejecting almost false-positive calls were determined. First-tier variants were filtered using the following criteria: coverage depth ≥20×, mutant reads ≥8, and variant frequency ≥2%. Second-tier variants were filtered according to the following criteria: coverage depth ≥20×, mutant reads ≥10, and variant frequency ≥5% [31].
In DFCI, C > T mutations consistent with a 20:1 single-strand bias were filtered out based on the read pair orientation to remove artifacts resulting from the hydrolytic deamination of cytosine to form uracil, specifically in formalin-fixed, paraffin-embedded samples. The MutSigCV suite of tools and manual curation was used to identify significantly mutated genes [27].
Microsatellite Instability Analysis
For TCGA PanCancer Atlas and MSKCC samples, the microsatellite status was assessed via MSIsensor, a computational algorithm that analyses sequencing reads at designated microsatellite regions in tumor-normal pairs reporting the percentage of unstable loci as a cumulative score [32]. MSI sensor scores ≥10 were defined as MSI-high (MSI-H), scores ≥3 and <10 as MSI-intermediate (MSI-I), and scores <3 as microsatellite stable (MSS) [33]. For DFCI samples, microsatellite status was analyzed using 10 microsatellite markers (D2S123, D5S346, D17S250, BAT25, BAT26, BAT40, D18S55, D18S56, D18S67, and D18S487) as previously described [27].
Estimation of TMB
TMB was estimated from TCGA PanCancer Atlas for KRAS mutation (n = 212), BRAF mutation (n = 57), and double mutation (n = 6) as the total number of mutations per sample/38 Mb. Furthermore, TMB was estimated from MSKCC for KRAS mutation (n = 470), BRAF mutation (n = 104), and double mutation (n = 17) as the total number of mutations per sample/1.22 Mb. The denominators 38 and 1.22 Mb represented the estimated length of human exome (38 Mb) reported in the TCGA database [34] and the estimated length of captured region (tumor DNA) of 468 cancer-related genes in the MSKCC database, respectively [35] The samples were classified as TMB-high if they had ≥12 mutations per megabase (mut/Mb), as previously described [36]. Additionally, the TMB of single-mutant and double-mutant CRC mutants from the two datasets were integrated (TCGA, MSKCC). Based on the integrated data hosted on TCGA and MSKCC, we compared the TMB in patients with KRAS-mutant (n = 682), BRAF-mutant (n = 161), and double-mutant (n = 23) tumors.
BRAF Classification
Amino-acid changes in BRAF in single-mutant and double-mutant cases (TCGA, MSKCC, and DFCI) were classified into classes 1, 2, and 3 according to previous reports [8,9]. Amino-acid changes that did not belong to any of these classes were classified as unknown.
Comparison of Clinicopathological Features, MSI Status, and TMB of CRC Mutants in TCGA, MSKCC, and DFCI Datasets
We integrated the clinicopathological information of the CRC mutants from the three datasets (TCGA, MSKCC, and DFCI) and performed a comparative analysis among KRAS-mutant, BRAF-mutant, and double-mutant CRCs. In the DFCI dataset, data on histological type and TMB were not available. Therefore, histological type and TMB were measured only in TCGA and MSKCC datasets. The histological type information was not available for the DFCI dataset; therefore, the percentage for histological type was calculated from 543 cases in KRAS-mutant CRC, 126 cases in BRAF-mutant CRC, and 18 cases in double-mutant CRC. MSI status was calculated only in the TCGA and MSKCC datasets because the evaluation method was different in the DFCI dataset. Conversely, information on CIMP was only available in the DFCI dataset. Instances of N/A were omitted from the percentage calculation.
Statistical Analyses
The clinicopathological features of patients with KRAS and BRAF single-mutant and double-mutant CRC were analyzed using the chi-square and Fisher’s exact tests. Comparisons between the single mutation (KRAS or BRAF) and double mutations in hotspot and other mutation sites of KRAS and V600E and non-V600E mutations of BRAF were analyzed using the chi-square test. The TMB of patients with KRAS mutant, BRAF mutant, and double-mutant CRC was analyzed using the Mann–Whitney U test. The Bonferroni post-test correction was used to reduce the likelihood of false positives. Between-group comparisons (KRAS mutation vs. double mutation, BRAF mutation vs. double mutation) were performed, and p < 0.025 (0.05/2) was considered statistically significant. All statistical analyses were performed using R software, version 4.0.3 (R Foundation for Statistical Computing, Vienna, Austria).
Results
Comparison of the Frequency of the Double KRAS/BRAF Mutation Between the Present and Previous Studies
The data from previous reports and the present study are summarized in Tables 1, 2. In our study, the double KRAS/BRAF mutation frequency from the integrated analysis of TCGA, MSKCC, and DFCI data was 1.2% (29/2,347). The frequency was 1% (6/594) in TCGA, 1.5% (17/1,134) in MSKCC, and 1% (6/619) in DFCI data. Codons 12 (exon 2), 13 (exon 2), 59 (exon 3), 61 (exon 3), 117 (exon 4), and 146 (exon 4) are the hotspots of KRAS mutation [37]. Codon 600 (exon 15) is the hotspot of V600E and non-V600E BRAF mutations [3,37] The numbers of each of the three mutations (KRAS mutation, BRAF mutation, and double mutation) that occurred in the hotspots of codons 12, 13, 61, 117, and 146 in TCGA, MSKCC, and DFCI were determined.
TABLE 1
| References | KRAS mut (%) | BRAF mut (%) | Double mut (%) | Sequence area |
|---|---|---|---|---|
| 13 | 397/1,063 (37.4) | 60/999 (6.9) | 1/999 (0.1) | KRAS (codon 12,13)BRAF (V600E) |
| 14 | 450/1,077 (41.8) | 26/397 (6.5) | 0/397 (0) | KRAS (codon 12, 13)BRAF (V600E) |
| 15 | 90/315 (28.8) | 33/315 (10.6) | 0/315 (0) | KRAS (codon 12, 13)BRAF (V600E) |
| 16 | 565/1,294 (43.7) | 102/1,189 (8.5) | 0/1,189 (0) | KRAS (codon 12, 13, 61)BRAF (codon 600) |
| 17 | 63/200 (31.5) | 14/200 (6.5) | 0/200 (0) | KRAS (codon 12, 13)BRAF (codon 15, V600) |
| 18 | 136/315 (43.2) | 11/309 (3.6) | 0/309 (0) | KRAS (codon 12, 13)BRAF (V600E) |
| 19 | 299/747 (40.0) | 36/761 (4.7) | 0/761 (0) | KRAS (codon 12, 13, 61, 146)BRAF (V600E) |
| Total | 2000/5,011 (39.9) | 282/4,170 (6.8) | 1/4,170 (0.02) |
Frequency of KRAS mutation, BRAF mutation, and KRAS/BRAF double mutation and target sites reported in previous studies.
Mut, mutation.
TABLE 2
| Data set | KRAS mut (%) | BRAF mut (%) | Double mut (%) | Method | Sequence area |
|---|---|---|---|---|---|
| TCGA | 212/594 (35.7) | 57/594 (9.6) | 6/594 (1.0) | NGS | Whole exon |
| MSKCC | 470/1,134 (41.4) | 104/1,134 (9.2) | 17/1,134 (1.5) | NGS | CDS of 468 genes, including KRAS, BRAF |
| DFCI | 167/619 (27) | 121/619 (19.5) | 6/619 (1.0) | NGS | Whole exon |
| Total | 849/2347 (36.1) | 282/2347 (12) | 29/2347 (1.2) |
Frequency of KRAS mutation, BRAF mutation, and KRAS/BRAF double mutation and the sequence area obtained from the integrated analysis of TCGA, MSKCC, and DFCI datasets in this study.
CDS, coding sequence; mut, mutation; mut, mutation; NGS, next-generation sequence.
Comparison of the Hotspots of Single and Double Mutations of KRAS and BRAF
Double-mutant CRC cases had significantly more non-hotspot and non-V600E mutations than single-mutant CRC cases (p < 0.01, respectively). The KRAS single mutation appeared in 97.8% hotspots, whereas the double mutation appeared in 68.8% hotspots and 31.3% other sites (Figure 1A). Moreover, 79.5% of the BRAF single mutations were of the V600E type, whereas 20.5% of them were of the non-V600E type. Although 22.2% of the BRAF mutations in double-mutant CRC were of the V600E type, 77.8% were of the non-V600E type (Figure 1B).
FIGURE 1
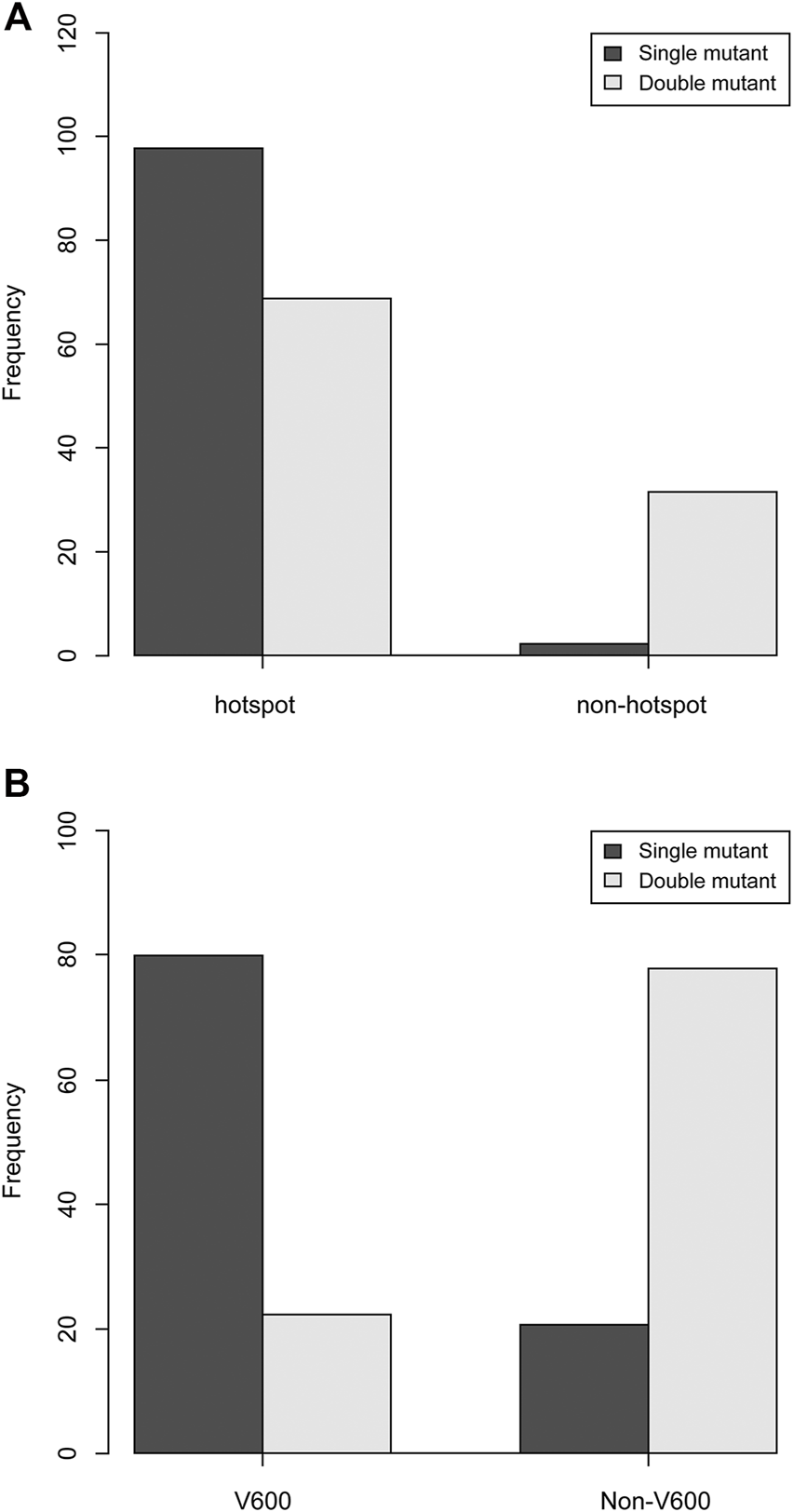
Comparison of hotspots of KRAS and BRAF single and double mutations. (A) Compared to single-mutant CRC, double-mutant CRC had significantly fewer hotspot mutations and had more non-hotspot mutations (p < 0.01). (B) Compared to single-mutant CRC, double-mutant CRC had significantly fewer V600 mutations and had more non-V600 mutations (p < 0.01).
Clinicopathological Features of Patients With Double-Mutant CRC in Cohort Data Associated With TCGA, MSKCC, and DFCI Datasets
In total, 2,347 CRC samples were identified in the cohorts associated with TCGA, MSKCC, and DFCI datasets. The clinicopathological characteristics, MSI status, and TMB for patients with double-mutant CRC are summarized in Table 3. The clinicopathological features of patients with double-mutant CRC, namely age (average), sex, tumor site, histological type, tumor grade, and stage data, were obtained from the cBioPortal. MSI status and TMB were classified as described in materials and methods. However, some patients, for whom the data of histological type and MSI status were unavailable (indicated by N/A in Table 3), were omitted from the percentage calculation. Mutations were identified more in males, and the occurrence of tumor sites was slightly higher in the right side of the colon. Regarding the histopathological type, the conventional type was the most common; however, mucinous and poor differentiation were also observed. Histological grades G2 and G3 were observed in most cases, whereas G1 was absent. With respect to the MSI status, 52.4 and 46.6% of the cases were classified as MSS and MSI cases, respectively (MSI-I, MSI-H). TMB-low and TMB-high were observed in 39.1 and 60.1% of the cases, respectively. CIMP-low and -high accounted for 60% and 40% of the cases, respectively. Regarding BRAF classification, 22.2% of double-mutant CRC cases were class 1, 0% were class 2, 16.7% were class 3, and 61.1% were unknown.
TABLE 3
| Characteristics | Categories | TCGA (n = 6) | MSKCC (n = 17) | DFCI (n = 6) | Total |
|---|---|---|---|---|---|
| Age (average) | x | 46–84 (71.7) | 24–78 (50.8) | 61–86 (71.8) | 24–84 (59.5) |
| Sex (%) | Male/Female | 6 (100)/0 (0) | 12 (70.6)/5 (29.4) | 4 (66.7)/2 (33.3) | 22 (75.9)/7 (24.1) |
| Tumor site (%) | Left/Right | 2 (33.3)/4 (66.7) | 8 (47.1)/9 (52.9) | 2 (33.3)/4 (66.7) | 12 (41.4)/17 (58.6) |
| Histological type (%) | Conventional | 5 (83.3) | 3 (25.0) | N/A | 8 (44.4) |
| Conventional with mucinous | 0 (0) | 3 (25.0) | N/A | 3 (16.7) | |
| Mucinous | 1 (16.7) | 2 (16.7) | N/A | 3 (16.7) | |
| PDC | 0 (0) | 4 (33.3) | N/A | 4 (22.2) | |
| N/A | 0 | 5 | 6 | 11 | |
| histological characteristics (%) | |||||
| Tumor grade (%) | G1/G2/G3 | 0 (0)/3 (50)/3 (50) | 0 (0)/7 (58.3)/5 (41.7) | 0 (0)/5 (83.3)/1 (16.7) | 0 (0)/15 (62.5)/9 (37.5) |
| N/A | 0 | 5 | 0 | 5 | |
| Stage (%) | Ⅰ/Ⅱ/Ⅲ/Ⅳ | 1 (16.7)/4 (66.7)/1 (16.7)/0 (0) | 0 (0)/5 (29.4)/5 (29.4)/7 (41.2) | 2 (33.3)/3 (33.3)/1 (16.7)/0 (0) | 3 (10.3)/12 (41.4)/7 (24.1)/7 (24.1) |
| MSI status (%) | MSS/MSI-I/MSI-H | 2 (33.3)/0 (0)/4 (66.7) | 10 (58.8)/2 (11.8)/5 (29.4) | 3 (75)/0 (0)/2 (25) | 12 (52.4)/2 (8.7)/9 (39.1) |
| N/A | 0 | 0 | 1 | 1 | |
| TMB (%) | TMB-low (<12 mut/Mb) | 1 (16.7) | 8 (47.1) | N/A | 9 (39.1) |
| TMB-high (≥12 Mb) | 5 (83.3) | 9 (52.9) | N/A | 14 (60.9) | |
| CIMP | CIMP-low/CIMP-high | N/A | N/A | 3 (60)/2 (40) | 3 (60)/2 (40) |
| N/A | 6 | 17 | 1 | 24 | |
| BRAF class | |||||
| 0 | 2 | 8 (22.2) | |||
| 2 | 0 | 0 (0) | |||
| 3 | 1 | 4 | 1 | 6 (16.7) | |
| unknown | 6 | 13 | 3 | 22 (61.1) |
Clinicopathological information regarding KRAS/BRAF double-mutant CRC (n = 29).
Mut, mutation, PDC, poorly differentiated adenocarcinoma; MSS; microsatellite stability; MSI-I; Microsatellite instability-intermediate; MSI-H; Microsatellite instability-high; TMB, tumor mutation burden; CIMP, CpG island methylator phenotype.
Comparison of Clinicopathological Features, MSI Status, and TMB Among Patients With KRAS-Mutant, BRAF-Mutant, and Double-Mutant CRC Based on Integrated Analysis of Information Available in TCGA, MSKCC, and DFCI Datasets
The comparison among KRAS-mutant, BRAF-mutant, and double-mutant tumors in three datasets is summarized in Table 4. Double-mutant tumors were observed predominantly in males, and their frequency (75.9%) was significantly higher than that of the KRAS- and BRAF-mutant tumors (47.9 and 36.5%, respectively; p < 0.01) in males. Histological types of the double-mutant cases were significantly different from those observed in KRAS-mutant cases (p = 0.02); however, the difference between BRAF- and double-mutant cases was not significant (p = 0.59). Similarly, the histological grades differed significantly between KRAS- and double-mutant cases (p < 0.01), although not between BRAF- and double-mutant cases (p = 0.86). MSI status of double-mutant cases significantly differed from those observed in KRAS- and BRAF-mutant cases (p < 0.01 and p = 0.02, respectively). Contrarily, for CIMP, no significant difference among the three groups was observed. The mean TMB in KRAS-mutant CRC was 10.8 mut/Mb (median = 5.0), whereas that in BRAF-mutant CRC and double-mutant CRC was 24.7 mut/Mb (median = 8.2 mut/Mb) and 59.4 mut/Mb (median = 36.1), respectively. The TMB in double-mutant CRC was significantly higher than that in KRAS-mutant CRC (p < 0.01, Figure 2A) but was not significantly higher than that in BRAF-mutant CRC (p < 0.026, Figure 2B). TMB was frequently high in patients with double-mutant CRC compared to that in patients with KRAS-mutant tumors. However, the frequency of TMB did not differ among patients with BRAF- and double-mutant tumors (p = 0.026). Significantly fewer cases of double-mutant CRC were classified as BRAF class 1 and more were classified as unknown (p < 0.01) (Figure 3).
TABLE 4
| Characteristics | Categories | KRAS mut (n = 849) | BRAF mut (n = 283) | Double mut (n = 29) | p-value (KRAS vs double) | p-value (BRAF vs double) |
|---|---|---|---|---|---|---|
| Frequency | 36.2% | 12.0% | 1.2% | |||
| Age (average) | 20–93 (61.3) | 26–90 (66.5) | 24–86 (59.5) | |||
| Sex (%) | Male | 406 (47.9) | 103 (36.5) | 22 (75.9) | <0.01 | <0.01 |
| Female | 442 (52.1) | 179 (63.5) | 7 (24.1) | |||
| N/A | 1 | 0 | 0 | |||
| Site (%) | Left | 436 (53) | 70 (25.3) | 12 (41.4) | 0.26 | 0.08 |
| Right | 387 (47) | 207 (74.7) | 17 (58.6) | |||
| N/A | 26 | 5 | 0 | |||
| Histological type (%) | Conventional | 405 (74.6) | 71 (56.3) | 8 (44.4) | 0.02 | 0.59 |
| Conventional with mucinous | 51 (9.4) | 17 (13.5) | 3 (16.7) | |||
| Mucinous | 52 (9.6) | 23 (18.3) | 3 (16.7) | |||
| PDC | 32 (5.9) | 13 (10.3) | 4 (22.2) | |||
| Signet | 2 (0.4) | 0 (0) | 0 (0) | |||
| MANEC | 1 (0.2) | 1 (0.8) | 0 (0) | |||
| Medullary | 0 (0) | 1 (0.8) | 0 (0) | |||
| N/A | 139 | 35 | 5 | |||
| Grade (%) | G1 | 25 (3.7) | 2 (0.9) | 0 (0) | <0.01 | 0.86 |
| G2 | 554 (81.1) | 146 (63.8) | 15 (62.5) | |||
| G3 | 104 (15.2) | 81 (35.4) | 9 (37.5) | |||
| N/A | 166 | 53 | 5 | |||
| Stage (%) | Ⅰ | 88 (10.7) | 37 (13.3) | 3 (10.3) | 0.07 | 0.88 |
| Ⅱ | 174 (21.1) | 96 (34.4) | 12 (41.4) | |||
| Ⅲ | 214 (26) | 64 (22.9) | 7 (24.1) | |||
| Ⅳ | 347 (42.7) | 82 (29.4) | 7 (24.1) | |||
| N/A | 26 | 3 | 0 | |||
| MSI status (%) | MSS | 615 (90.7) | 100 (62.5) | 12 (52.2) | <0.01 | 0.02 |
| MSI-I | 13 (1.9) | 0 (0) | 2 (8.7) | |||
| MSI-H | 54 (7.9) | 60 (37.5) | 9 (39.1) | |||
| N/A | 0 | 1 | 0 | |||
| CIMP | CIMP-low | 117 (90.7) | 29 (30.5) | 3 (60) | 0.08 | 0.32 |
| CIMP-high | 12 (9.3) | 66 (69.5) | 2 (40) | |||
| N/A | 38 | 26 | ||||
| BRAF class | <0.01 | |||||
| 1 | N/A | 225 (79.5) | 8 (22.2) | |||
| 2 | N/A | 12 (4.2) | 0 (0) | |||
| 3 | N/A | 22 (7.8) | 6 (16.7) | |||
| unknown | N/A | 24 (8.5) | 22 (61.1) | |||
| TMB (mut/Mb) (%) | TMB-low | 619 (90.8) | 92 (57.1) | 9 (39.1) | <0.01 | 0.026 |
| TMB-high | 63 (9.2) | 69 (42.9) | 14 (60.9) |
Comparison of clinicopathological information among KRAS-mutant CRC; BRAF-mutant CRC; and double-mutant CRC obtained by integrating the information available in TCGA; MSKCC; and DFCI datasets (n = 2347).
Mut, mutation; PDC, poorly differentiated adenocarcinoma; MANEC, mixed adenoneuroendocrine carcinoma; MSS; microsatellite stability; MSI-I; Microsatellite instability-intermediate; MSI-H; Microsatellite instability-high; TMB, Tumor mutation burden. p-values were calculated by Fisher’s exact test.
FIGURE 2
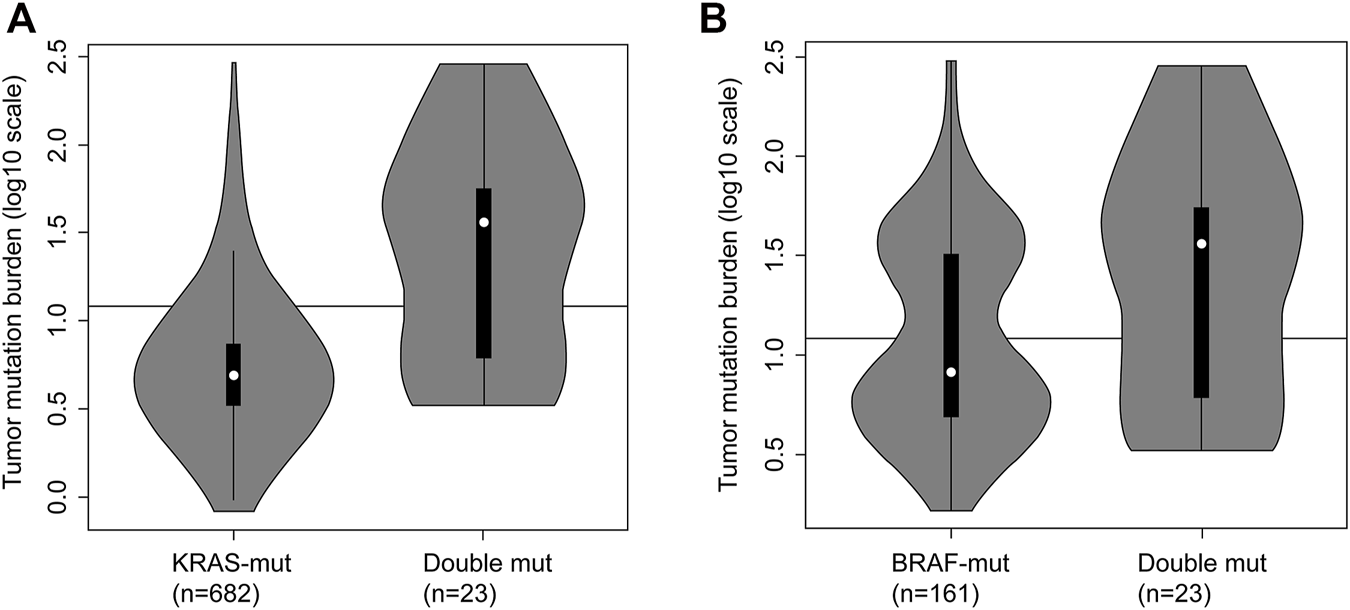
Tumor mutation burden (TMB) in KRAS-mutant CRC, BRAF-mutant CRC and double-mutant CRC. (A) TMB in KRAS-mutant CRC and double-mutant CRC. Comparison of TMB in patients with KRAS-mutant (n = 682) and double-mutant (n = 23) tumors based on integrated data hosted on The Cancer Genome Atlas (TCGA) and Memorial Sloan-Kettering Cancer Center (MSKCC; Mann–Whitney U test, p < 0.01). Black line indicating 12 mut/Mb represents the threshold for TMB-high. For KRAS-mutant CRC, the frequency of TMB-high was 9.2% (63/682); for double-mutant CRC, the frequency was 60.9% (14/23). (B) Tumor mutation burden in BRAF-mutant CRC and double-mutant CRC. Comparison of TMB in patients with BRAF-mutant (n = 161) and double-mutant (n = 23) tumors based on integrated data hosted on TCGA and MSKCC (the Mann–Whitney U test, p = 0.026). Black line indicating 12 mut/Mb represents the threshold for TMB-high. For BRAF-mutant CRC, the frequency of TMB-high was 42.9% (69/161); for double-mutant CRC, the frequency was 60.9% (14/23).
FIGURE 3
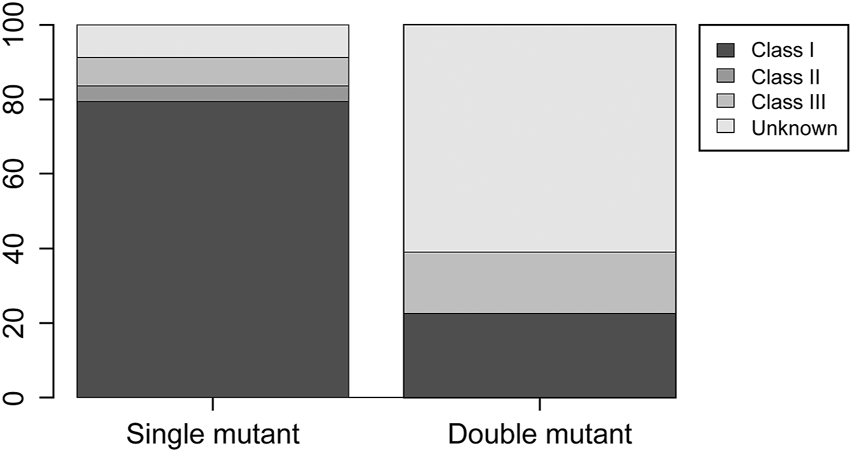
Distribution of BRAF class 1, 2, and 3 and unknown in BRAF single-mutant CRC and double-mutant CRC cases. Compared to single-mutant CRC, double-mutant CRC had significantly fewer BRAF class 1 and more unknown (p < 0.01).
Discussion
In this study, the integrated results of three datasets from cBioPortal indicate that the frequency of the double-mutant CRC is 1.2%, which is greater than that of previous reports (0.02%) [13–19], This difference can be primarily attributed to the difference in sequencing methods used in the present versus previous studies. Most previous studies have reported the mutations only in hotspots, such as codons 12 and 13 of KRAS and codon 600 (i.e., V600E) of BRAF. However, in this study, we analyzed whole exome sequence (WES) and coding sequence (CDS) datasets of KRAS and BRAF. We inferred that the KRAS and BRAF mutations identified from the hotspots were mutually exclusive, as reported in several previous studies. The double mutations tended to occur at a relatively higher frequency outside hotspots (Figures 1A,B).
Double-mutant CRC demonstrated slightly higher occurrence in the right side of the colon and displayed mucinous differentiation and poor differentiation significantly more often than KRAS-mutant CRC. G3 was significantly more frequent than KRAS-mutant (Table 4). Double-mutant CRC demonstrated significantly more MSI-H than KRAS-mutant CRC (Table 4). Considering the clinicopathological features, several double-mutant CRCs differed significantly from KRAS-mutant CRC, although they displayed similar characteristics with BRAF-mutant CRC. Regarding TMB, double-mutant CRC demonstrated the highest TMB-high ratio, which significantly exceeded that of KRAS-mutant CRC (Table 4). These findings demonstrated that double-mutant CRC displayed a higher TMB value than those reported earlier [38]. To the best of our knowledge, this is the first study to analyze the three datasets collectively, identify the double mutations in CRC, and assess the clinicopathological features, MSI status, and TMB of KRAS/BRAF double-mutant CRC and compare them with KRAS and BRAF single mutation CRC.
Previous studies have identified the benefits of using EGFR inhibitors (i.e., cetuximab) for treating KRAS- and BRAF-mutant CRCs [39,40]. Tumor biology and drug sensitivity change with the site of the KRAS [41] and BRAF mutations. The drug sensitivity of non-V600 BRAF remains controversial, and there are several unclear points as discussed below [8]. Recent studies have reported that non-V600 BRAF mutations are associated with low response rates to EGFR inhibitors in CRC [42,43]. However, there have also been reports of patients with class 3 BRAF mutations who responded to EGFR inhibitors and chemotherapy [19]. In the current study, there were significantly more non-V600 BRAF mutations in the double-mutant CRC cases than in the single-mutant CRC cases. However, 61.1% of double-mutant CRC cases were classified as unknown. From these results, it appears that double-mutant CRC may have different biology compared to single-mutant CRC. Currently, no effective treatment has been established for double-mutant CRC. Attempts to treat double-mutant CRC by chemotherapy with FOLFOX (fluorouracil + folinic acid + oxaliplatin) have been presented in several case reports. Since the effect of EGFR could not be observed secondary to KRAS and BRAF mutations, all patients had received FOLFOX (fluorouracil + folinic acid + oxaliplatin) [21–25]. However, none of them exhibited a significant effect, and five of the seven patients died. From the results of this study, it can be observed that detection of double-mutant CRC is dependent on sequencing methods. As panel sequencing and whole-exome or -genome sequencing by next-generation sequencing has recently become widespread in clinical settings, double-mutant CRC may be detected more frequently. Further studies are necessary to modify and develop new chemotherapy regimens by including immune checkpoint inhibitors to achieve disease control in patients with KRAS/BRAF double-mutant CRC.
The study had certain limitations. First, the percentages calculated in this study might not be accurate, as we used different datasets, and the data for some of the tested characteristics were not available (N/A) or some categories had several instances of N/As. Second, the effect of data analysis methods that might incur false positives and false negatives and affect the overall frequency estimation was not evaluated in this study. Therefore, examining studies reporting the positive and negative false positives to gain insight into the influence of the data analysis methods in determining the frequency of double mutations could be interesting and useful. Third, we did not analyze the NRAS-mutant CRC, a biomarker for anti-EGFR treatment, in addition to KRAS and BRAF mutations. It has been presented that NRAS mutations are rare CRCs and do not appear to be associated with any of the molecular features, including mutation of KRAS, BRAF, PIK3CA, MSI, and CIMP [44]. Moreover, the frequency of double mutations involving NRAS mutations is rare [45,46]. Only three samples displayed triple mutations from the cases studied here (n = 2347), including NRAS, and analysis was impossible. Therefore, screening NRAS single and double mutants using a larger dataset might contribute to the development of an effective treatment strategy. Fourth, we could not carry out survival analysis because the stage and treatment methods for analyzing prognosis were not stringently standardized. Therefore, more data are required to determine whether the KRAS/BRAF double mutation can serve as a prognostic factor.
Conclusion
We demonstrated that the occurrence frequency of the KRAS/BRAF double-mutant CRC was higher than that reported previously, suggesting that using a larger sample size and improved technologies that cover the sequencing information of WES and CDS datasets of cancer-related genes will be efficient in identifying the rare double mutations at a higher rate. Moreover, the findings suggest that double-mutant CRC is characterized by a higher occurrence in men and slight right-sided predominance. Pathologically, there were characterized by a significantly higher incidence of mucinous differentiation, poor differentiation, and a high histological grade (G3) than that of KRAS-mutant CRC. At the molecular level, significantly more MSI-high and higher TMB values were observed compared with those of KRAS-mutant CRC.
Statements
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
Experiment design, SH and TS; Data analysis, SU and TK; Statistical analyses, TK. All authors were involved in the writing of the paper and approved the submitted manuscript.
Acknowledgments
The Bioinformatics Consultation Forum, The Society for Biotechnology, Japan, taught the authors how to use the web tools (cBioPortal and GDC Data Portal) for bioinformatics analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.por-journal.com/articles/10.3389/pore.2022.1610206/full#supplementary-material
Supplementary Table S1Summary of the legacy identifier (COSMIC), pathogenicity, and allele frequency of the KRAS and BRAF mutations of double-mutant CRC.
Abbreviations
BRAF, v-RAF murine sarcoma viral oncogene homolog B1; CIMP, CpG island methylator phenotype; CDS, coding sequence; CRC, colorectal cancer; DFCI, Dana-Farber Cancer Institute; EGFR, epidermal growth factor receptor; KRAS, Kirsten rat sarcoma viral oncogene homolog; MSI, microsatellite instability; MSKCC, Memorial Sloan-Kettering Cancer Center; MSS, microsatellite stable; TCGA, The Cancer Genome Atlas; TMB, tumor mutation burden; WES, whole exome sequence.
References
1.
WHO Classification of Tumours Editorial Board. WHO Classification of Tumours In: Digestive System Tumours. 5th ed. Lyon: IARC (2019).
2.
Cancer Genome Atlas Network. Comprehensive Molecular Characterization of Human colon and Rectal Cancer. Nature (2012) 487:330–7. 10.1038/nature11252
3.
Morkel M Riemer P Bläker H Sers C . Similar but Different: Distinct Roles for KRAS and BRAF Oncogenes in Colorectal Cancer Development and Therapy Resistance. Oncotarget (2015) 6:20785–800. 10.18632/oncotarget.4750
4.
Lin JS Webber EM Senger CA Holmes RS Whitlock EP . Systematic Review of Pharmacogenetic Testing for Predicting Clinical Benefit to Anti-EGFR Therapy in Metastatic Colorectal Cancer. Am J Cancer Res (2011) 1:650–62.
5.
Castellano E Downward J . RAS Interaction with PI3K: More Than Just Another Effector Pathway. Genes & Cancer (2011) 2:261–74. 10.1177/1947601911408079
6.
Eklöf V Wikberg M.L Edin S Dahlin A.M Jonsson B-A Öberg A et al The Prognostic Role of KRAS, BRAF, PIK3CA and PTEN in Colorectal Cancer. Br J Cancer (2013) 108:2153–63. 10.1038/bjc.2013.212
7.
Afrăsânie V.-A Marinca MV Alexa-Stratulat T Gafton B Păduraru M Adavidoaiei AM et al KRAS, NRAS, BRAF, HER2 and Microsatellite Instability in Metastatic Colorectal Cancer - Practical Implications for the Clinician. Radiol Oncol (2019) 53:265–74. 10.2478/raon-2019-0033
8.
Dankner M Rose AAN Rajkumar S Siegel PM Watson IR Classifying BRAF Alterations in Cancer: New Rational Therapeutic Strategies for Actionable Mutations. Oncogene (2018) 37:3183–99. 10.1038/s41388-018-0171-x
9.
Yao Z Yaeger R Rodrik-Outmezguine VS Tao A Torres NM Chang MT et al Tumours with Class 3 BRAF Mutants Are Sensitive to the Inhibition of Activated RAS. Nature (2017) 548:234–8. 10.1038/nature23291
10.
Yoshino T Arnold D Taniguchi H Pentheroudakis G Yamazaki K Xu R-H et al Pan-Asian Adapted ESMO Consensus Guidelines for the Management of Patients with Metastatic Colorectal Cancer: A JSMO-ESMO Initiative Endorsed by CSCO, KACO, MOS, SSO and TOS. Ann Oncol (2018) 29:44–70. 10.1093/annonc/mdx738
11.
Rajagopalan H Bardelli A Lengauer C Kinzler KW Vogelstein B Velculescu VE RAF/RAS Oncogenes and Mismatch-Repair Status. Nature (2002) 418:934. 10.1038/418934a
12.
Nakayama I Hirota T Shinozaki E BRAF Mutation in Colorectal Cancers: From Prognostic Marker to Targetable Mutation. Cancers (2020) 12:3236. 10.3390/cancers12113236
13.
Van Cutsem E Köhne C-H Láng I Folprecht G Nowacki MP Cascinu S et al Cetuximab Plus Irinotecan, Fluorouracil, and Leucovorin as First-Line Treatment for Metastatic Colorectal Cancer: Updated Analysis of Overall Survival According to Tumor KRAS and BRAF Mutation Status. Jco (2011) 29:2011–9. 10.1200/JCO.2010.33.5091
14.
Lamy A Blanchard F Le Pessot F Sesboüé R Di Fiore F Bossut J et al Metastatic Colorectal Cancer KRAS Genotyping in Routine Practice: Results and Pitfalls. Mod Pathol (2011) 24:1090–100. 10.1038/modpathol.2011.60
15.
Price TJ Hardingham JE Lee CK Weickhardt A Townsend AR Wrin JW et al Impact of KRAS and BRAF Gene Mutation Status on Outcomes from the Phase III AGITG MAX Trial of Capecitabine Alone or in Combination with Bevacizumab and Mitomycin in Advanced Colorectal Cancer. Jco (2011) 29:2675–82. 10.1200/JCO.2010.34.5520
16.
Maughan TS Adams RA Smith CG Meade AM Seymour MT Wilson RH et al Addition of Cetuximab to Oxaliplatin-Based First-Line Combination Chemotherapy for Treatment of Advanced Colorectal Cancer: Results of the Randomised Phase 3 MRC COIN Trial. The Lancet (2011) 377:2103–14. 10.1016/S0140-6736(11)60613-2
17.
Wang X Lu YY An YX Wang X Zhao QC KRAS, BRAF and PIK3CA Mutations in Human Colorectal Cancer: Relationship with Metastatic Colorectal Cancer. Oncol Rep (2011) 25:1691–7. 10.3892/or.2011.1217
18.
Bokemeyer C Bondarenko I Hartmann JT de Braud F Schuch G Zubel A et al Efficacy According to Biomarker Status of Cetuximab Plus FOLFOX-4 as First-Line Treatment for Metastatic Colorectal Cancer: The OPUS Study. Ann Oncol (2011) 22:1535–46. 10.1093/annonc/mdq632
19.
De Roock W Claes B Bernasconi D De Schutter J Biesmans B Fountzilas G et al Effects of KRAS, BRAF, NRAS, and PIK3CA Mutations on the Efficacy of Cetuximab Plus Chemotherapy in Chemotherapy-Refractory Metastatic Colorectal Cancer: a Retrospective Consortium Analysis. Lancet Oncol (2010) 11:753–62. 10.1016/S1470-2045(10)70130-3
20.
Sahin IH Kazmi SMA Yorio JT Bhadkamkar NA KeeGarrett BKCR Garrett CR Rare Though Not Mutually Exclusive: A Report of Three Cases of Concomitant KRAS and BRAF Mutation and a Review of the Literature. J Cancer (2013) 4:320–2. 10.7150/jca.3619
21.
Larki P Gharib E Yaghoob Taleghani M Khorshidi F Nazemalhosseini-Mojarad E Asadzadeh Aghdaei H Coexistence of KRAS and BRAF Mutations in Colorectal Cancer: A Case Report Supporting the Concept of Tumoral Heterogeneity. Cell J (2017) 19:113–7. 10.22074/cellj.2017.5123
22.
Vittal A Middinti A Kasi Loknath Kumar A Are All Mutations the Same? A Rare Case Report of Coexisting Mutually Exclusive KRAS and BRAF Mutations in a Patient with Metastatic colon Adenocarcinoma. Case Rep Oncological Med (2017) 2017:1–3. 10.1155/2017/2321052
23.
Deshwar A Margonis GA Andreatos N Barbon C Wang J Buettner S et al Double KRAS and BRAF Mutations in Surgically Treated Colorectal Cancer Liver Metastases: An International, Multi-Institutional Case Series. Ar (2018) 38:2891–5. 10.21873/anticanres.12535
24.
Midthun L Shaheen S Deisch J Senthil M Tsai J Hsueh C-T Concomitant KRAS and BRAF Mutations in Colorectal Cancer. J Gastrointest Oncol (2019) 10:577–81. 10.21037/jgo.2019.01.10
25.
Cafiero C Re A D’Amato G Surico PL Surico G Pirrelli M et al KRAS and BRAF Concomitant Mutations in a Patient with Metastatic colon Adenocarcinoma: An Interesting Case Report. Case Rep Oncol (2020) 13:595–600. 10.1159/000507882
26.
Yaeger R Chatila WK Lipsyc MD Hechtman JF Cercek A Sanchez-Vega F et al Clinical Sequencing Defines the Genomic Landscape of Metastatic Colorectal Cancer. Cancer Cell (2018) 33:125–36. 10.1016/j.ccell.2017.12.004
27.
Giannakis M Mu XJ Shukla SA Qian ZR Cohen O Nishihara R et al Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cel Rep (2016) 15:857–65. 10.1016/j.celrep.2016.03.075
28.
Tate JG Bamford S Jubb HC Sondka Z Beare DM Bindal N et al COSMIC: The Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res (2019) 47:D941–D947. 10.1093/nar/gky1015
29.
Cancer Genome Atlas Research Network. Integrated Genomic Analyses of Ovarian Carcinoma. Nature (2011) 474:609–15. 10.1038/nature10166
30.
Liu Y Sethi NS Hinoue T Schneider BG Cherniack AD Sanchez-Vega F et al Comparative Molecular Analysis of Gastrointestinal Adenocarcinomas. Cancer Cell (2018) 33:735–21. 10.1016/j.ccell.2018.03.010
31.
Cheng DT Mitchell TN Zehir A Shah RH Benayed R Syed A et al Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT). J Mol Diagn (2015) 17:251–64. 10.1016/j.jmoldx.2014.12.006
32.
Niu B Ye K Zhang Q Lu C Xie M McLellan MD et al MSIsensor: Microsatellite Instability Detection Using Paired Tumor-normal Sequence Data. Bioinformatics (2014) 30:1015–6. 10.1093/bioinformatics/btt755
33.
Latham A Srinivasan P Kemel Y Shia J Bandlamudi C Mandelker D et al Microsatellite Instability Is Associated with the Presence of Lynch Syndrome Pan-Cancer. Jco (2019) 37:286–95. 10.1200/JCO.18.00283
34.
Chalmers ZR Connelly CF Fabrizio D Gay L Ali SM Ennis R et al Analysis of 100,000 Human Cancer Genomes Reveals the Landscape of Tumor Mutational burden. Genome Med (2017) 9:34. 10.1186/s13073-017-0424-2
35.
Chan TA Yarchoan M Jaffee E Swanton C Quezada SA Stenzinger A et al Development of Tumor Mutation burden as an Immunotherapy Biomarker: Utility for the Oncology Clinic. Ann Oncol (2019) 30:44–56. 10.1093/annonc/mdy495
36.
Fabrizio DA George Jr TJ Jr Dunne RF Frampton G Sun J Gowen K et al Beyond Microsatellite Testing: Assessment of Tumor Mutational burden Identifies Subsets of Colorectal Cancer Who May Respond to Immune Checkpoint Inhibition. J Gastrointest Oncol (2018) 9:610–7. 10.21037/jgo.2018.05.06
37.
Gilson P Franczak C Dubouis L Husson M Rouyer M Demange J et al Evaluation of KRAS, NRAS and BRAF Hotspot Mutations Detection for Patients with Metastatic Colorectal Cancer Using Direct DNA Pipetting in a Fully-Automated Platform and Next-Generation Sequencing for Laboratory Workflow Optimisation. PLOS ONE (2019) 14:e0219204. 10.1371/journal.pone.0219204
38.
Wu H-X Wang Z-X Zhao Q Chen D-L He M-M Yang L-P et al Tumor Mutational and Indel burden: A Systematic Pan-Cancer Evaluation as Prognostic Biomarkers. Ann Transl Med (2019) 7:640. 10.21037/atm.2019.10.116
39.
Karapetis CS Khambata-Ford S Jonker DJ O'Callaghan CJ Tu D Tebbutt NC et al K-rasMutations and Benefit from Cetuximab in Advanced Colorectal Cancer. N Engl J Med (2008) 359:1757–65. 10.1056/NEJMoa0804385
40.
Van Cutsem E Köhne C-H Hitre E Zaluski J Chang Chien C-R Makhson A et al Cetuximab and Chemotherapy as Initial Treatment for Metastatic Colorectal Cancer. N Engl J Med (2009) 360:1408–17. 10.1056/NEJMoa0805019
41.
De Roock W Jonker DJ Di Nicolantonio F Sartore-Bianchi A Tu D Siena S et al Association of KRAS p.G13D Mutation with Outcome in Patients with Chemotherapy-Refractory Metastatic Colorectal Cancer Treated with Cetuximab. JAMA (2010) 304:1812–20. 10.1001/jama.2010.1535
42.
Shinozaki E Yoshino T Yamazaki K Muro K Yamaguchi K Nishina T et al Clinical Significance of BRAF Non-v600e Mutations on the Therapeutic Effects of Anti-EGFR Monoclonal Antibody Treatment in Patients with Pretreated Metastatic Colorectal Cancer: The Biomarker Research for Anti-EGFR Monoclonal Antibodies by Comprehensive Cancer Genomics (BREAC) Study. Br J Cancer (2017) 117:1450–8. 10.1038/bjc.2017.308
43.
Hsu H-C Thiam TK Lu Y-J Yeh CY Tsai W-S You JF et al Mutations of KRAS/NRAS/BRAF Predict Cetuximab Resistance in Metastatic Colorectal Cancer Patients. Oncotarget (2016) 7:22257–70. 10.18632/oncotarget.8076
44.
Irahara N Baba Y Nosho K Shima K Yan L Dias-Santagata D et al NRAS Mutations Are Rare in Colorectal Cancer. Diagn Mol Pathol (2010) 19:157–63. 10.1097/PDM.0b013e3181c93fd1
45.
Sanchez-Ibarra HE Jiang X Gallegos-Gonzalez EY Cavazos-González AC Chen Y Morcos F et al KRAS, NRAS, and BRAF Mutation Prevalence, Clinicopathological Association, and Their Application in a Predictive Model in Mexican Patients with Metastatic Colorectal Cancer: A Retrospective Cohort Study. PLOS ONE (2020) 15:e0235490. 10.1371/journal.pone.0235490
46.
Vittal A Sharma D Samanta I Kasi A Rare Case of Triple Mutant (KRAS + NRAS + BRAF) Metastatic colon Adenocarcinoma. BMJ Case Rep (2019) 12:e221816. 10.1136/bcr-2017-221816
Summary
Keywords
cancer, KRAS , BRAF , bioinformatic analysis, colon, double mutation
Citation
Uchida S, Kojima T and Sugino T (2022) Frequency and Clinicopathological Characteristics of Patients With KRAS/BRAF Double-Mutant Colorectal Cancer: An In Silico Study. Pathol. Oncol. Res. 28:1610206. doi: 10.3389/pore.2022.1610206
Received
22 November 2021
Accepted
26 January 2022
Published
24 February 2022
Volume
28 - 2022
Edited by
Andrea Ladányi, National Institute of Oncology (NIO), Hungary
Updates
Copyright
© 2022 Uchida, Kojima and Sugino.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shiro Uchida, Dr.Uchida@gmail.com, orcid.org/0000-0002-7086-896X
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.