Abstract
SLC13A4 is a sodium sulfate co-transporter, which is expressed in brains, placentas, thymes and other tissues, plays an essential role in maintaining the metabolic balance of sulfate in vivo. The TCGA database shows that it is differentially expressed in a variety of tumors, but its prognostic value in tumors has not been clarified. TCGA, Oncomine and Timer databases were used to analyze SLC13A4 mRNA expression in cancer tissues and normal tissues, and its correlation with clinical prognosis in head and neck tumor. The CIBERSORT database was used to analyze the correlation between SLC13A4 expression and the infiltration of immune cells. SLC13A4 enrichment analysis was carried out by GSEA. SLC13A4 mRNA levels were significantly lower in head and neck tumors than in paracancer tissues. SLC13A4 expression in Head and neck squamous cell carcinoma (HNSCC) was closely related to tumor pathological grade and clinical stage. Decreased SLC13A4 expression was associated with poor overall survival (OS), progression free survival (PFS), disease specific survival (DSS) and recurrence free survival (RFS) in HNSCC patients. The expression of SLC13A4 was negatively correlated with Monocytes, M1 macrophages, M2 macrophages, resting CD4+ memory T cells, resting NK cells and activated NK cells, but positively correlated with neutrophils, plasma cells, T follicular helper cells, gamma delta T cells, regulatory T cells and naive B cells. In addition, the genes in SLC13A4 low-expression group were mainly concentrated in immunity-related activities, viral diseases, typical tumor pathways and metabolism. The SLC13A4 high expression group was mainly enriched in metabolic pathways. These suggest that SLC13A4 may be a potential prognostic biomarker in HNSC and correlated with immune infiltrates.
Introduction
Most head and neck cancers are squamous cell carcinomas that originate in the upper respiratory epithelia (located in oral cavity, oropharynx, larynx or hypopharynx). Head and neck squamous cell carcinoma (HNSCC) tend to develop in the mucous membrane of the mouth, nose, and throat. It is the sixth most common cancer around the globe, with a 5-year mortality rate of 50% [1,2]. Although the application of various treatment modalities, and combination use of surgical, radiotherapeutic and chemotherapeutic treatments can significantly reduce the short-term and long-term survival of the malignancy, the situation remains grim [3]. The immune cells and stromal infiltration contribute to the tumor microenvironment (TME), in which various immune cells act as either promoters or inhibitors of cancers, and they are closely related to the prognosis of patients [4]. Intensive researches demonstrated that PD-1 inhibitors exert a significant therapeutic effect on HNSCC [5,6]. In the clinical practice, early diagnosis and accurate prognostic prediction of HNSCC patient remain a challenge due to the lack of specific biomarkers. To improve the long-term survival of tumor patients, it is of great importance to identify the biomarkers that are conductive to early diagnosis, precise outcome prediction of cancers and to the identification of people who well respond to immunotherapy.
A feature shared by all members of solute carrier family 13 (SLC13) is the multi-bonded proteins encoding 8–13 transmembrane domains, and is found in a wide array of tissues. SLC13 family has 5 members. In terms of gene codes and function, they fall into two categories: sodium sulfate co-transporter (SLC13A1, SLC13A4) and sodium carboxylic acid co-transporter (SLC13A2, SLC13A3, SLC13A5) [7]. SLC13A4 is one of the major transporters of sulfate metabolism and plays an anti-apoptotic part by mediating the transmembrane transportion of thiosulfate on cell membrane, regulating the concentration of intracellular sulfate and modulate caspase-3 sulfurization [8]. SLC13A4 is very different from other sulfate transporters, and its functions have been preliminarily explored in tonsil, placenta, testis and central nervous system, but the physiological implication of its low-level expression in heart, thymus and liver has yet to be elucidated [9–12]. Moreover, its roles in tumors remains poorly understood. The mutated sulfate transporter gene and/or disordered sulfate metabolism are closely related to multiple human diseases, such as aberrant early embryonic development, chondrodysplasias and Hunter syndrome, which may be the predisposing factors of neurodevelopmental diseases, such as mental retardation and autism spectrum disorder [11,13–15]. Thiosulfate is one of the important inorganic sulfates in the body and the fourth most abundant anion in mammalian plasma, and is crucial for a great many physiological functions [16,17]. In prostate cancer, the content of thiosulfate is used as an auxiliary diagnostic marker [18]. All these highlight the important role that sulfate transporter protein plays in human physiology. In addition, thiosulfate is an important product of H2S metabolism. H2S was reported to induce DNA damage and affect the cell signal transduction and cell cycle, and participate in a variety of physiological and pathological processes, including tumorigenesis and development [19–21]. These further indicate that the sulfate process of metabolic pathway may be closely involved in TME.
This study examined the differential expression, clinical relevance and prognostic value of SLC13A4 in HNSCC by searching RNA-seq and clinical data in the TCGA public database. On the basis of its expression in thymus, we further analyzed the potential pathways and possible molecular mechanisms by which SLC13A4 is involved in HNSCC tumor immune infiltration by searching cibersort and GSEA database.
Materials and Methods
TCGA Database
All level 3 RNA expression data (Workflow Type: HTSeq-FPKM) from 502 HNSCC cancer tissues and 44 adjacent-cancer tissues were retrieved from the Cancer Genome Atlas (TCGA) database (https://tcga-data.nci.nih.gov/tcga/) [22]. The clinical information of HNSCC patients, including age at diagnosis, gender, HPV status, radiation therapy, pathological stage, T stage, N stage, M stage, grades, overall survival time, overall survival status, disease free survival time, disease free survival status, disease specific survival time, disease specific survival status, progression-free survival time, progression-free survival status were also downloaded from the TCGA. TCGA-HNSCC has 500 HNSCC patients with clinical information, but the information for each patient is not complete. In detail, TCGA-HNSCC provides gender information for 500 patients, age information for 499 patients, HPV status information for 481 patients, T stage information for 444 patients, N stage information for 407 patients, and M stage information for 187 patients, pathological stage information for 432 patients, grade information for 481 patients and radiation therapy information for 429 patients. Furthermore, 500 patients’ overall survival status information, 499 patients’ overall survival time information, progression free survival time and status, 474 patients’ disease specific survival time and status, 125 patients’ disease specific survival time and status were provided. All data are included in the study. According to the publication guidelines, datasets can be used for publication without restrictions or limitations.
GEO Database
RNA-seq data of SLC13A4 in GSE2837, GSE27020 and GSE41613 were obtained from Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) [23] to study the relationship between SLC13A4 expression level and prognosis of HNSCC patients. Normalized expression matrix files and sequencing platform annotations of the gene sets were downloaded.
Oncomine Database Analysis
Oncomine database (https://www.oncomine.org/resource/main.html) [24] was used for analyzing SLC13A4 expression in various cancer types. The cut-off value of p value and fold change were 0.01 and 2, respectively.
TIMER Database Analysis
TIMER database (https://cistrome.shinyapps.io/timer/) [25] contained 10,897 samples, including 32 different kinds of cancer types covered by the TCGA dataset. We used the database to evaluate the SLC13A4 expression in tumors of various types and its correlation with T cell exhaustion markers.
The Human Protein Atlas
The human protein atlas (HPA) contains the detailed RNA sequences of 37 major normal tissue types and immunohistochemical results of tissue microarray involving 44 different tissue types. HPA (https://www.proteinatlas.org) [26] has the data about the expression and localization of human proteins in tissues and organs. All the data are openly accessible. We analyzed the expression of SLC13A4 protein in normal oral tissues and oral squamous cell carcinoma by on the basis of immunohistochemistry image in HPA. The antibody used is HPA048582 (SIGMA-ALDRICH).
Survival Analysis
Survival analysis was performed by using survival and survminer packages in R. 499 patients had detailed survival time data and progression-free survival time data, 474 patients had disease specific survival time data, and 124 patients had disease free survival time data in TCGA. 97 Patients had overall survival time data in GSE41613. 28 Patients had recurrence-free survival time data in GSE2837. 109 Patients had disease specific survival time data in GSE27020. All the time data were employed for survival analysis. The median value of survival time from TCGA database and mean value of survival time from GEO database was used as the cut-off value for the survival analysis. The survival curve was plotted by using Kaplan-Meier method, and log rank was used for significance assessment. A p < 0.05 was considered statistically significant.
Immune Infiltration Analysis
To estimate the abundance profile of the tumor-infiltrating immune cells (TIICs) in HNSCC, CIBERSORT (http://cibersort.stanford.edu/) [26] computational method was applied for analyzing mRNA sequence data of all tumor samples. Based on the validated leukocyte gene signature matrix (termed LM22), the proportion of infiltration of 22 kinds of immune cells in each sample was statistically analyzed. In each sample, the ratio of all TIICs was equal to 1.
Barplot, Corrplot, Vioplot and Scatter Plot
Barplot and corrplot were visualized by using R software with package corrplot, while vioplot was produced with packages BiocManager and vioplot. Except for package BiocManager, scatter plotting was performed by employing R software with packages ggplot2, ggpubr and ggExtra.
Gene Set Enrichment Analysis
GSEA is supported by the Broad Institute Website (http://software.broadinstitute.org/gsea/index.jsp) [27]. We got HNSCC RNA-sequencing data from TCGA, and performed gene set enrichment analysis by using GSEA software (V4.0.2). We downloaded kegg, hallmark and immune signatures gene sets from Molecular Signatures Database to investigate the possible biological functions of SLC13A4. Gene sets with nominal p value < 0.05 and FDR ≤ 25% were considered statistically significant.
Statistical Analysis
All statistical analyses were conducted by using SPSS 26.0 statistical software (SPSS, Chicago, IL), GraphPad prism 7.0, R X64 4.0.0 and Bioconductor packages (https://www.bioconductor.org/). Wilcoxon test was used to compare the different proportions of TIICs between high SLC13A4 expression group and low expression group. Fisher test was employed to analyze the relationship between SLC13A4 mRNA expression and clinical characteristics. Univariate Cox regression models were utilized to study the prognostic value of clinicopathological parameters for HNSCC. The multivariate Cox regression models were used to assess the association between SLC13A4 mRNA expression and survival adjusted for age, gender, T stage, N stage, pathological stage, grade, HPV status and radiation therapy. Overall survival (OS), disease free survival (DFS), progression free survival (PFS), disease specific survival (DSS) and recurrence free survival (RFS) curves were calculated by Kaplan-Meier method. GraphPad Prism 7.0 was applied for statistical analysis of clinical information. Normal distribution data were subjected to parametric test, and non-normal distribution data to Mann-Whitney test. Significant difference was set at p < 0.05.
Results
Expression of SLC13A4 in Various Human Cancer Types
Using Oncomine database, we analyzed SLC13A4 gene expression in different tumors and the adjacent tissues. The results are shown in Figure 1A. SLC13A4 expression was higher in lymphoma tissues than in adjacent tissues, and was lower in esophageal cancer, HNSCC and sarcoma. To further evaluate SLC13A4 expression in various tumors, we used Timer database to analyze the transcriptional sequence data of 22 TCGA tumors. The results are shown in Figures 1B,C. SLC13A4 expression was lower in breast cancer, head and neck squamous cell carcinoma, kidney chromophobe, thyroid cancer, and higher in renal clear cell carcinoma, hepatocellular carcinoma, cholangiocarcinoma, and rectal adenocarcinoma. In addition, immunohistochemical analyses available from the HPA are shown in Figure 1D, SLC13A4 expression was lower in tumor tissues when compared to normal tissues.
FIGURE 1
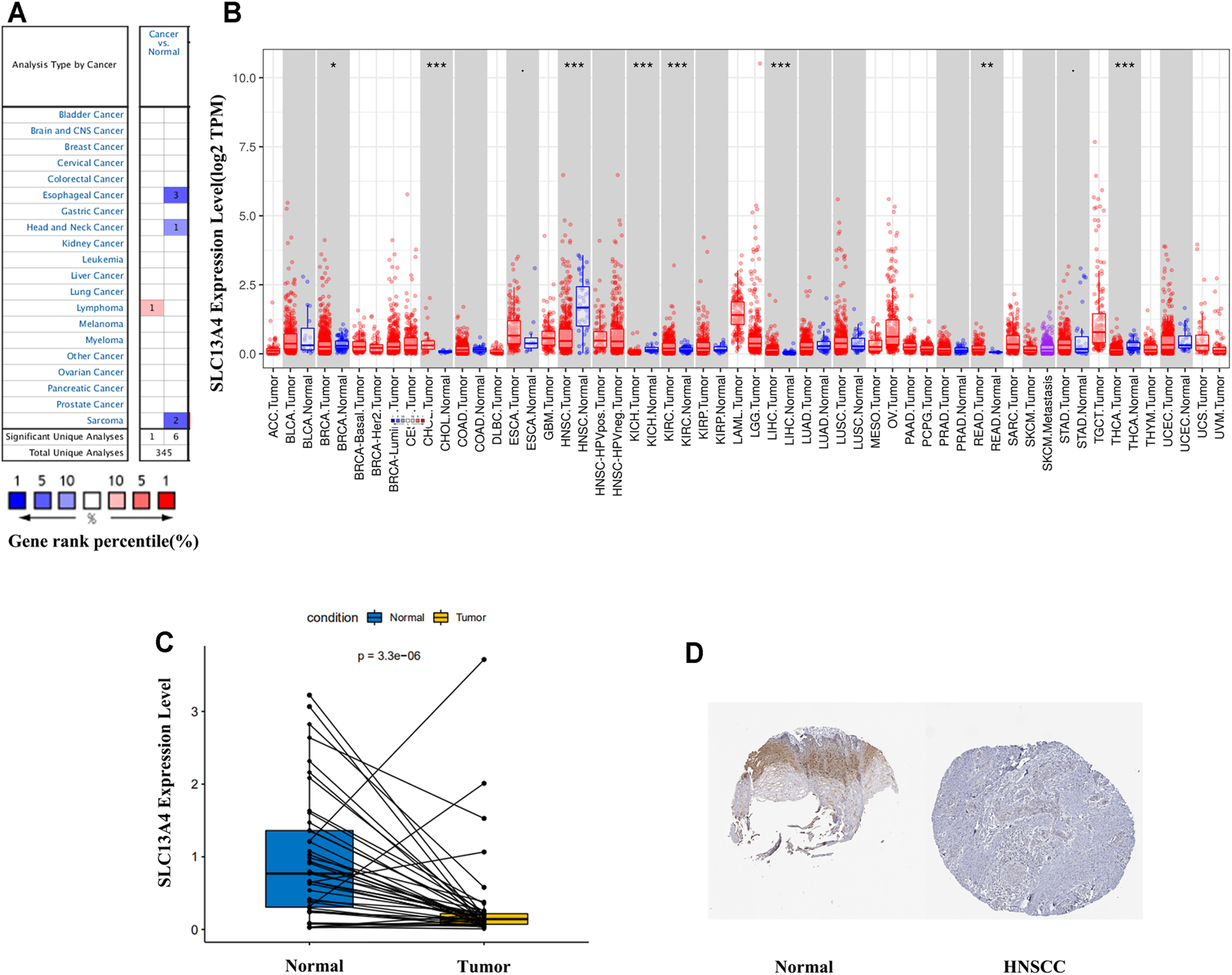
SLC13A4 expression levels in different types of human cancers. (A) Increased or decreased SLC13A4 in data sets of different cancers compared with normal tissues in the Oncomine database. (B) Human SLC13A4 expression levels in different tumor types from TCGA database were determined by TIMER (*p < 0.05, **p < 0.01, ***p < 0.001). (C) SLC13A4 expression levels between adjacent and tumor tissues in HNSCC patients from TCGA database. (D) The expression of SLC13A4 protein in normal oral tissues and oral squamous cell carcinoma was observed using immunohistochemistry via HPA.
Correlation Between SLC13A4 Expression and Clinical Features in HNSCC Patients
We obtained the data on transcription sequences and clinical characteristics of HNSCC patients from the TCGA database, and divided HNSCC patients into the high-expression group and the low-expression group in terms of the median mRNA expression of SLC13A4. The results are shown in Table 1. HPV status and overall survival in HNSCC patients were significantly correlated with SLC13A4 mRNA expression. HPV-positivity rate (17.7%) was higher in high SLC13A4 expression group than in its low-expression counterpart (10.9%). For the overall survival status, the proportion of deaths in HNSCC patients with high SLC13A4 expression (34.4%) was significantly lower than that with low SLC13A4 expression (52.4%).
TABLE 1
| Characters | Level | Low expression of SLC13A4 | High expression of SLC13A4 | p Value |
| n | 250 | 250 | ||
| Gender | Female | 68 (27.2%) | 65 (26.0%) | 0.840 |
| Male | 182 (72.8%) | 185 (74.0%) | ||
| Age | <60 years | 100 (40.2%) | 120 (48.0%) | 0.087 |
| ≥60 years | 149 (59.8%) | 130 (52.0%) | ||
| HPV status | HPV(−) | 212 (89.1%) | 200 (82.3%) | 0.038 |
| HPV(+) | 26 (10.9%) | 43 (17.7%) | ||
| T Stage (%) | T1 | 19 (8.6%) | 26 (11.6%) | 0.252 |
| T2 | 67 (30.5%) | 65 (29.0%) | ||
| T3 | 55 (25.0%) | 41 (18.3%) | ||
| T4 | 79 (35.9%) | 92 (41.1%) | ||
| N Stage (%) | N0 | 72 (36.7%) | 99 (46.9%) | 0.192 |
| N1 | 34 (17.3%) | 31 (14.7%) | ||
| N2 | 87 (44.4%) | 77 (36.5%) | ||
| N3 | 3 (1.5%) | 4 (1.9%) | ||
| M Stage (%) | M0 | 80 (100.0%) | 106 (99.1%) | 0.572 |
| M1 | 0 (0.0%) | 1 (0.9%) | ||
| Pathological stage (%) | Stage I | 11 (5.1%) | 14 (6.5%) | 0.921 |
| Stage II | 36 (16.7%) | 34 (15.7%) | ||
| Stage III | 40 (18.6%) | 38 (17.5%) | ||
| Stage IV | 128 (59.5%) | 131 (60.4%) | ||
| Grade | G1 | 23 (9.5%) | 38 (15.9%) | 0.069 |
| G2 | 155 (64.0%) | 144 (60.3%) | ||
| G3 | 64 (26.4%) | 55 (23.0%) | ||
| G4 | 0 (0.0%) | 2 (0.8%) | ||
| Radiation therapy (%) | No | 76 (36.2%) | 74 (33.8%) | 0.614 |
| Yes | 134 (63.8%) | 145 (66.2%) | ||
| Overall survival_status | Alive | 119 (47.6%) | 164 (65.6%) | <0.001 |
| Dead | 131 (52.4%) | 86 (34.4%) |
Relationship between clinical character and SLC13A4 expression in HNSCC.
We grouped the clinical characteristics of the patients and compared the differences in mRNA expression of SLC13A4 in each group. The results are shown in Figure 2 and Table 1. SLC13A4 expression showed statistically significant differences among T stage, N stage and pathological grades, exhibiting an overall tendency that the more malignant the tumor, the lower the mRNA expression of SLC13A4. The mRNA expression level of SLC13A4 at T1 stage was significantly higher than those at T2 stage (p = 0.0342) and T3 stage (p = 0.0175). SLC13A4 expression level at N0 stage was significantly higher than that at N2 stage (p = 0.0022). SLC13A4 expression at grade 1 was significantly higher than those at grade 2 (p = 0.0287) and grade 3 (p = 0.003). Since there were only two samples that were in grade 4, no statistical comparison was made.
FIGURE 2
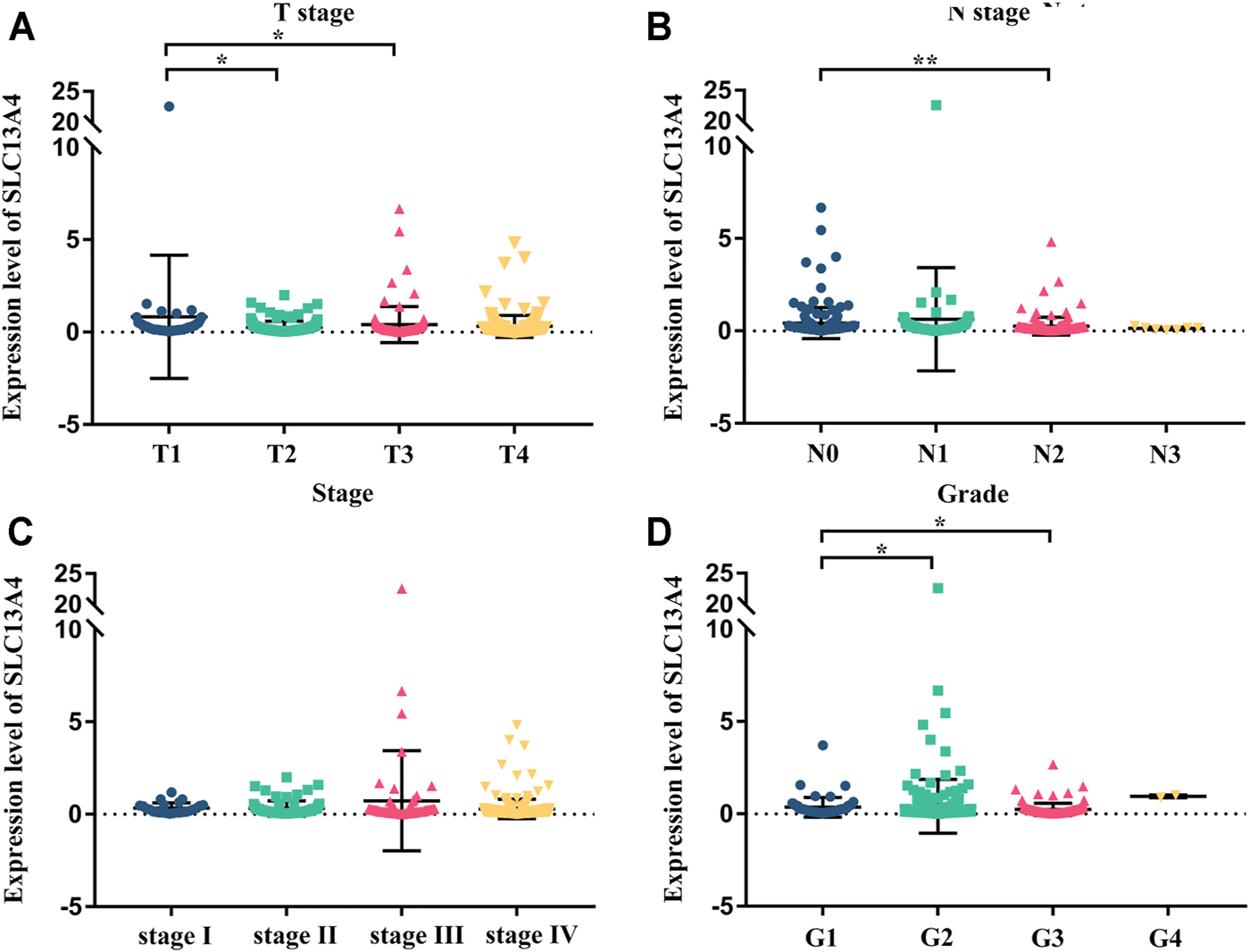
Relationship between mRNA expression of SLC13A4 and clinical characteristic in HNSCC patients. mRNA expression of SLC13A4 were remarkably correlative with T stage, N stage and grade. Patients in advanced T stage (A), N stage (B), clinical stage (C) and grade (D) tends to show lower mRNA expression of SLC13A4 (*p < 0.05, **p < 0.01).
Univariate and Multivariate Cox Regression Analysis of Factors Affecting Survival in HNSCC Patients
To assess the risk factors influencing the prognosis of HNSCC patients, Cox regression models were used to analyze the clinical data of the patients. Univariate analysis revealed that SLC13A4 expression level, T Stage, N Stage and pathological stage are high risk factors affecting overall survival, disease specific survival and progression free survival in patients with HNSCC. In addition, HPV negative status is also a higher risk factor affecting both overall survival and progression free survival, and radiation therapy is a risk factor only affecting overall survival (Table 2). Moreover, after adjusted for age, gender, T Stage, N Stage, pathological stage, grade, HPV status and radiation therapy, the relationship between SLC13A4 mRNA expression and overall survival, disease specific survival and progression free survival were still significant (Tables 3–5).
TABLE 2
| Characters | Overall survival | Progression free survival | Disease specific survival | |||
|---|---|---|---|---|---|---|
| HR (95%CI) | p Value | HR (95%CI) | p value | HR (95%CI) | p Value | |
| Gender | 0.751 (0.564–1.001) | 0.051 | 1.042 (0.753–1.443) | 0.802 | 0.956 (0.643–1.421) | 0.825 |
| Age | 1.293 (0.982–1.703) | 0.067 | 1.105 (0.830–1.472) | 0.493 | 1.075 (0.757–1.525) | 0.688 |
| HPV status | 0.503 (0.306–0.826) | 0.007 | 0.598 (0.372–0.961) | 0.034 | 0.569 (0.314–1.032) | 0.063 |
| T Stage | 1.300 (1.126–1.501) | <0.001 | 1.286 (1.105–1.495) | 0.001 | 1.374 (1.137–1.662) | 0.001 |
| N Stage | 1.532 (1.295–1.813) | <0.001 | 1.433 (1.205–1.703) | <0.001 | 1.729 (1.384–2.160) | <0.001 |
| Pathological Stage | 1.416 (1.182–1.696) | <0.001 | 1.339 (1.114–1.608) | 0.002 | 1.519 (1.187–1.944) | 0.001 |
| Grade | 1.103 (0.895–1.358) | 0.358 | 1.075 (0.860–1.344) | 0.526 | 1.144 (0.870–1.503) | 0.336 |
| Radiation Therapy | 0.639 (0.467–0.874) | 0.005 | 0.893 (0.640–1.248) | 0.508 | 0.780 (0.512–1.190) | 0.250 |
| SLC13A4 | 0.601 (0.457–0.789) | <0.001 | 0.587 (0.440–0.784) | <0.001 | 0.461 (0.320–0.664) | <0.001 |
Univariate Cox regression analysis of factors affecting survival in HNSCC patients.
TABLE 3
| Variables | Model 1 (n = 499)a | Model 2 (n = 499)b | Model 3 (n = 386)c | Model 4 (n = 331)d | ||||
|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | p Value | HR (95% CI) | p Value | HR (95% CI) | p Value | HR (95% CI) | p Value | |
| SLC13A4* | ||||||||
| Low expression group | 1.665 (1.267–2.187) | p < 0.001 | 1.640 (1.248–2.156) | p < 0.001 | 1.774 (1.279–2.461) | 0.001 | 1.571 (1.072–2.301) | 0.020 |
| High expression group | Reference | Reference | Reference | Reference | Reference | Reference | Reference | Reference |
| Age | ||||||||
| <60 years | 0.814 (0.616–1.077) | 0.149 | 0.734 (0.524–1.029) | 0.073 | 0.846 (0.566–1.266) | 0.416 | ||
| ≥60 years | Reference | Reference | Reference | Reference | Reference | Reference | ||
| Gender | ||||||||
| Female | 1.250 (0.935–1.673) | 0.133 | 1.294 (0.912–1.837) | 0.149 | 1.400 (0.937–2.091) | 0.100 | ||
| Male | Reference | Reference | Reference | Reference | Reference | Reference | ||
| Grade | ||||||||
| G1 | 128.800 (0−1.355E+32) | 0.890 | 48.545 (0−3.450E+35) | 0.922 | ||||
| G2 | 179.261 (0−1.883E+32) | 0.883 | 54.690 (0−3.881E+35) | 0.920 | ||||
| G3 | 163.841 (0−1.722E+32) | 0.885 | 53.358 (0−3.787E+35) | 0.920 | ||||
| G4 | Reference | Reference | Reference | Reference | ||||
| Stage | ||||||||
| Stage I | 0.649 (0.113–3.736) | 0.628 | 0.402 (0.065–2.498) | 0.328 | ||||
| Stage II | 0.624 (0.232–1.678) | 0.350 | 0.357 (0.113–1.126) | 0.079 | ||||
| Stage III | 1.314 (0.653–2.643) | 0.444 | 0.728 (0.318–1.667) | 0.453 | ||||
| Stage IV | Reference | Reference | Reference | Reference | ||||
| T | ||||||||
| T1 | 0.627 (0.179–2.194) | 0.465 | 0.656 (0.178–2.412) | 0.526 | ||||
| T2 | 0.673 (0.368–1.232) | 0.199 | 0.726 (0.351–1.501) | 0.387 | ||||
| T3 | 0.936 (0.597–1.466) | 0.772 | 0.835 (0.490–1.422) | 0.506 | ||||
| T4 | Reference | Reference | Reference | Reference | ||||
| N | ||||||||
| N0 | 0.274 (0.103–0.731) | 0.010 | 0.347 (0.045–2.694) | 0.311 | ||||
| N1 | 0.229 (0.077–0.68) | 0.008 | 0.329 (0.040–2.707) | 0.301 | ||||
| N2 | 0.518 (0.207–1.299) | 0.161 | 0.735 (0.099–5.452) | 0.763 | ||||
| N3 | Reference | Reference | Reference | Reference | ||||
| HPV Status | ||||||||
| No | 2.227 (1.003–4.942) | 0.049 | ||||||
| Yes | Reference | Reference | ||||||
| Radiation therapy | ||||||||
| No | 2.958 (1.912–4.577) | p < 0.001 | ||||||
| Yes | Reference | Reference | ||||||
Relationship between SLC13A4 expression, clinical character and overall survival in HNSCC patients.
*The transcription level of SLC13A4 in HNSCC patients, and those above the median value were defined as the high expression group, while those below the median value were defined as the low expression group.
aUnadjusted.
bAdjusted for age and gender.
cAdjusted for age, gender, grade, stage, T stage and N stage.
dAdjusted for age, gender, grade, stage, T stage, N stage, HPV status and radiation therapy.
TABLE 4
| Variables | Model 1 (n = 499)a | Model 2 (n = 499)b | Model 3 (n = 386)c | Model 4 (n = 331)d | ||||
|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | p Value | HR (95% CI) | p Value | HR (95% CI) | p Value | HR (95% CI) | p Value | |
| SLC13A4* | ||||||||
| Low expression group | 1.702 (1.275–2.273) | p < 0.001 | 1.702 (1.275–2.273) | p < 0.001 | 1.94 (1.386–2.716) | p < 0.001 | 1.738 (1.195–2.526) | 0.004 |
| High expression group | Reference | Reference | Reference | Reference | Reference | Reference | Reference | Reference |
| Age | ||||||||
| <60 years | 0.908 (0.679–1.214) | 0.514 | 0.817 (0.581–1.149) | 0.246 | 0.756 (0.510–1.119) | 0.162 | ||
| ≥60 years | Reference | Reference | Reference | Reference | Reference | Reference | ||
| Gender | ||||||||
| Female | 0.92 (0.662–1.279) | 0.621 | 0.963 (0.658–1.41) | 0.848 | 0.970 (0.638–1.473) | 0.886 | ||
| Male | Reference | Reference | Reference | Reference | Reference | Reference | ||
| Grade | ||||||||
| G1 | 1.755 (0.303–10.176) | 0.531 | 1.361 (0.227–8.149) | 0.735 | ||||
| G2 | 0.686 (0.262–1.797) | 0.443 | 0.573 (0.196–1.672) | 0.308 | ||||
| G3 | 1.352 (0.646–2.828) | 0.423 | 1.030 (0.436–2.431) | 0.946 | ||||
| G4 | Reference | Reference | Reference | Reference | ||||
| Stage | ||||||||
| Stage I | 267.832 (0−2.816E+47) | 0.916 | 104.121 (0−1.053E+48) | 0.932 | ||||
| Stage II | 329.130 (0−3.457E+47) | 0.913 | 123.742 (0−1.25E+48) | 0.929 | ||||
| Stage III | 297.986 (0−3.132E+47) | 0.914 | 107.930 (0−1.091E+48) | 0.931 | ||||
| Stage IV | Reference | Reference | Reference | Reference | ||||
| T | ||||||||
| T1 | 0.438 (0.098–1.953) | 0.279 | 0.456 (0.101–2.065) | 0.308 | ||||
| T2 | 0.649 (0.356–1.183) | 0.158 | 0.691 (0.344–1.385) | 0.297 | ||||
| T3 | 0.805 (0.498–1.301) | 0.376 | 0.660 (0.380–1.148) | 0.142 | ||||
| T4 | Reference | Reference | Reference | Reference | ||||
| N | ||||||||
| N0 | 0.376 (0.128–1.103) | 0.075 | 0.586 (0.077–4.479) | 0.607 | ||||
| N1 | 0.252 (0.076–0.833) | 0.024 | 0.352 (0.042–2.915) | 0.333 | ||||
| N2 | 0.655 (0.237–1.807) | 0.414 | 1.077 (0.146–7.950) | 0.942 | ||||
| N3 | Reference | Reference | Reference | Reference | ||||
| HPV Status | ||||||||
| No | 2.155 (0.988–4.701) | 0.054 | ||||||
| Yes | Reference | Reference | ||||||
| Radiation therapy | ||||||||
| No | 1.471 (0.933–2.319) | 0.096 | ||||||
| Yes | Reference | Reference | ||||||
Relationship between SLC13A4 expression, clinical character and progression free survival in HNSCC patients
*The transcription level of SLC13A4 in HNSCC patients, and those above the median value were defined as the high expression group, while those below the median value were defined as the low expression group.
aUnadjusted.
bAdjusted for age and gender.
cAdjusted for age, gender, grade, stage, T stage and N stage.
dAdjusted for age, gender, grade, stage, T stage, N stage, HPV status and radiation therapy.
TABLE 5
| Variables | Model 1 (n = 474)a | Model 2 (n = 474)b | Model 3 (n = 371)c | Model 4 (n = 320)d | ||||
|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | p Value | HR (95% CI) | p Value | HR (95% CI) | p Value | HR (95% CI) | p Value | |
| SLC13A4* | ||||||||
| Low expression group | 2.171 (1.507–3.127) | p < 0.001 | 2.170 (1.505–3.128) | p < 0.001 | 2.527 (1.635–3.905) | p < 0.001 | 2.361 (1.411–3.951) | 0.001 |
| High expression group | Reference | Reference | Reference | Reference | Reference | Reference | Reference | Reference |
| Age | ||||||||
| <60 years | 0.948 (0.665–1.352) | 0.769 | 0.896 (0.587–1.368) | 0.611 | 0.877 (0.521–1.476) | 0.622 | ||
| ≥60 years | Reference | Reference | Reference | Reference | Reference | Reference | ||
| Gender | ||||||||
| Female | 0.983 (0.658–1.468) | 0.931 | 1.009 (0.629–1.619) | 0.970 | 1.049 (0.609–1.806) | 0.862 | ||
| Male | Reference | Reference | Reference | Reference | Reference | Reference | ||
| Grade | ||||||||
| G1 | 0.997 (0.073–13.683) | 0.998 | 0.647 (0.042–9.88) | 0.754 | ||||
| G2 | 0.580 (0.146–2.296) | 0.438 | 0.529 (0.107–2.614) | 0.435 | ||||
| G3 | 1.112 (0.443–2.792) | 0.821 | 0.595 (0.188–1.876) | 0.375 | ||||
| G4 | Reference | Reference | Reference | Reference | ||||
| Stage | ||||||||
| Stage I | 95.296 (0−3.502E+41) | 0.922 | 32.195 (0−3.166E+44) | 0.945 | ||||
| Stage II | 125.070 (0−4.585E+41) | 0.917 | 37.021 (0−3.633E+44) | 0.943 | ||||
| Stage III | 124.483 (0−4.567E+41) | 0.917 | 38.794 (0−3.807E+44) | 0.942 | ||||
| Stage IV | Reference | Reference | Reference | Reference | ||||
| T | ||||||||
| T1 | 0.461 (0.055–3.863) | 0.475 | 0.522 (0.058–4.724) | 0.563 | ||||
| T2 | 0.525 (0.240–1.150) | 0.107 | 0.466 (0.167–1.300) | 0.145 | ||||
| T3 | 0.991 (0.578–1.697) | 0.973 | 0.878 (0.461–1.67) | 0.691 | ||||
| T4 | Reference | Reference | Reference | Reference | ||||
| N | ||||||||
| N0 | 0.207 (0.066–0.647) | 0.007 | 0.257 (0.031–2.108) | 0.206 | ||||
| N1 | 0.178 (0.049–0.645) | 0.009 | 0.247 (0.028–2.215) | 0.212 | ||||
| N2 | 0.418 (0.149–1.175) | 0.098 | 0.600 (0.079–4.535) | 0.620 | ||||
| N3 | Reference | Reference | Reference | Reference | ||||
| HPV status | ||||||||
| No | 2.150 (0.759–6.09) | 0.150 | ||||||
| Yes | Reference | Reference | ||||||
| Radiation therapy | ||||||||
| No | 2.324 (1.295–4.17) | 0.005 | ||||||
| Yes | Reference | Reference | ||||||
Relationship between SLC13A4 expression, clinical character and disease specific survival in HNSCC patients.
*The transcription level of SLC13A4 in HNSCC patients, and those above the median value were defined as the high expression group, while those below the median value were defined as the low expression group.
aUnadjusted.
bAdjusted for age and gender.
cAdjusted for age, gender, grade, stage, T stage and N stage.
dAdjusted for age, gender, grade, stage, T stage, N stage, HPV status and radiation therapy.
Prognostic Value of SLC13A4 in HNSCC
We theorize that SLC13A4 expression is related to the prognosis of cancer patients. We used TCGA data to evaluate the effects of SLC13A4 expression on the total survival time, disease free survival time, disease specific survival time, and progression-free survival time of HNSCC patients. According to the median mRNA value of SLC13A4 expression, tumor samples were divided into a high expression group and a low expression group. Kaplan-Meier plot analysis was performed and results are shown in Figure 3. Compared with the low SLC13A4 expression group, the patients in the high-expression group had longer overall survival time, progression-free survival time and disease specific survival time (p < 0.001), and there was no significant difference in disease free survival time (p = 0.217). GEO validation cohort results are shown in Supplementary Figure S1. Compared with the low SLC13A4 expression group, the patients in the high-expression group had longer overall survival time (GSE41613, p = 0.010) and recurrence-free survival time (GSE2837, p = 0.011). Data from GSE27020 showed that there was no statistical difference in disease free survival time between SLC13A4 low expression group and SLC13A4 high expression group (p = 0.092). The findings suggest that SLC13A4 expression level is closely related to clinical characteristics and patients’ survival time, and is potentially of value in the assessment of the prognosis of HNSCC.
FIGURE 3
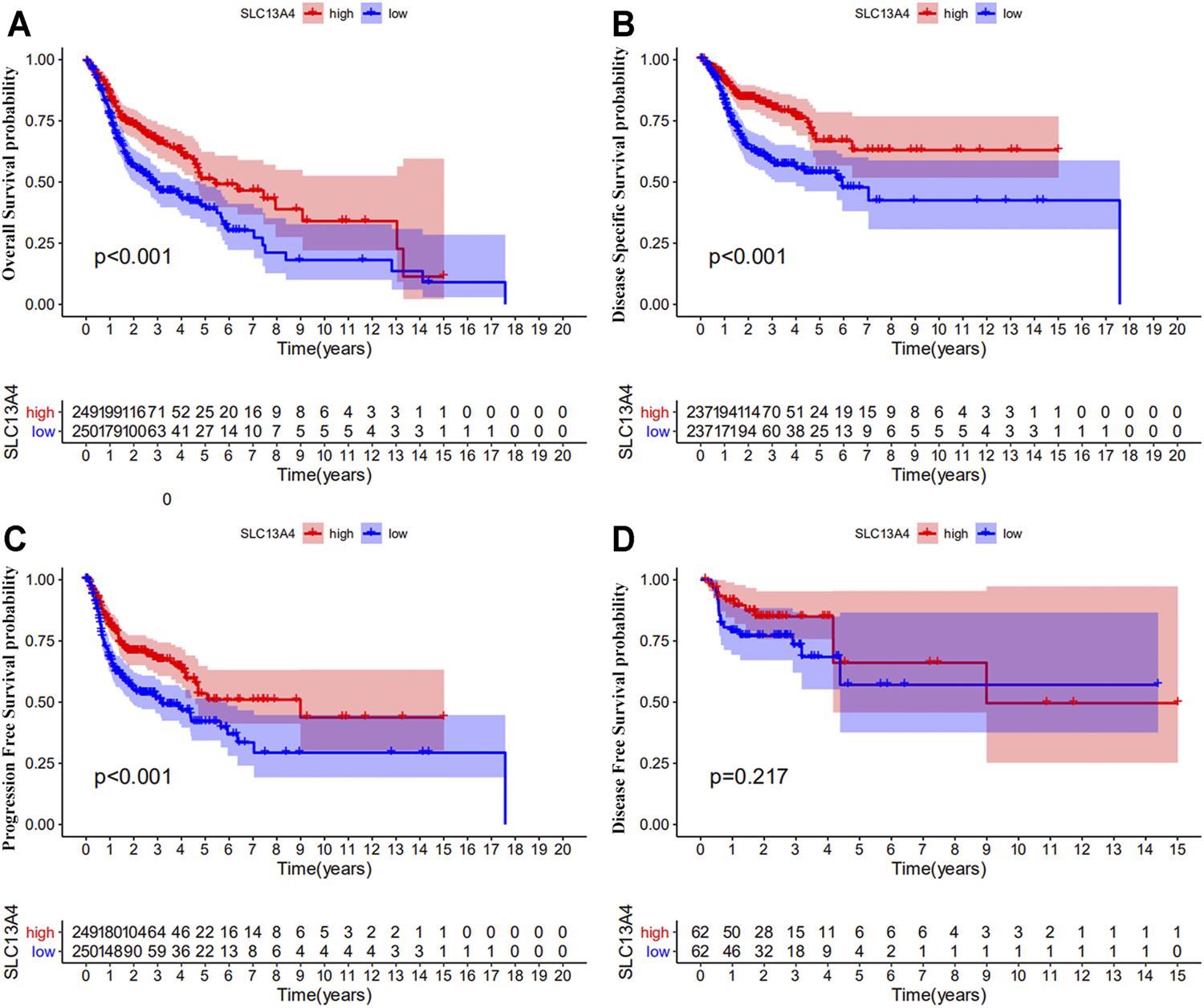
Prognostic significance of SLC13A4 in HNSCC. Kaplan-Meier survival curves of overall survival (A), disease-specific survival (B), progression free survival (C) and disease-free survival (D) between the high and low expression of SLC13A4 in HNSCC patients from TCGA database.
Correlation Between SLC13A4 Expression and Proportion of TIICs and Distribution of TIICs in HNSCC
Based on Cibersort algorithm, Barplot demonstrated the infiltration of 22 kinds of immune cells in HNSCC tumor samples. The Corrplot showed the correlation among 22 kinds of TIICs in the HNSCC samples. Tumor samples were divided into a high and a low expression group according to the median expression of SLC13A4. The results are shown in Figures 4, 5. The levels of naive B cells (p < 0.001), plasma cells (p < 0.001), T follicular helper cells ( p = 0.007), gamma delta T cells ( p = 0.001), regulatory T cells ( p < 0.01), neutrophils ( p = 0.011) were significantly higher in the high-expression group than in the low-expression group. The levels of resting CD4+ memory T cells ( p = 0.023), resting NK cells (p = 0.003), activated NK cells ( p = 0.007), M1 macrophages (p = 0.002), M2 macrophages ( p < 0.001)were significantly higher in the low-expression group than in the high-expression group. Correlation analysis showed that the expression of SLC13A4 was negatively correlated with monocytes, M1 macrophages, M2 macrophages, resting CD4+ memory T cells, resting NK cells and activated NK cells, but was positively correlated with neutrophils, plasma cells, T follicular helper cells, gamma delta T cells, regulatory T cells and naive B cells. Furthermore, the expression of SLC13A4 in HNSCC is negatively correlated with a variety of T cell exhaustion markers, including LAG3 ( p < 0.001), TIM-3( p < 0.001) and CTLA-4 ( p < 0.001), indicating its potential mechanism in regulating tumor immunity (Supplementary Table S1). These findings showed that SLC13A4 expression was related to the proportion of TIICs and distribution of TIICs in HNSCC, which further demonstrated that SLC13A4 expression affected the immune activity of HNSCC.
FIGURE 4
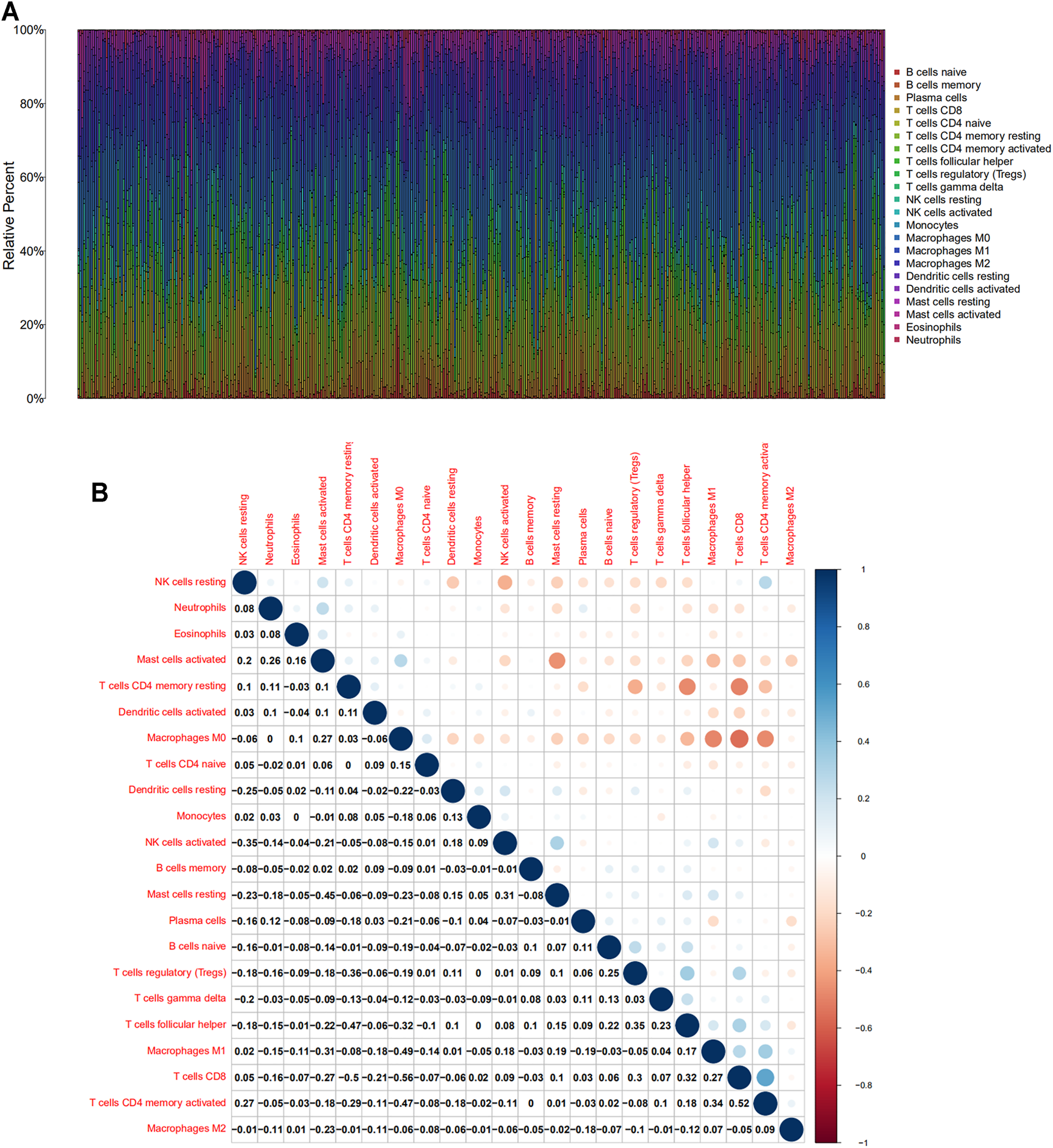
TIC profile and correlation analysis of HNSC samples. (A) Barplot showing the proportion of 22 kinds of TICs in HNSC tumor samples. (B) Corrplot representing the correlation between 22 kinds of TICs and quantity in each small box, indicating the p-value of cells that are correlated with each other. The depth of each tiny color box represents the corresponding correlation value between the two cells, and Pearson coefficient is used for significance test.
FIGURE 5
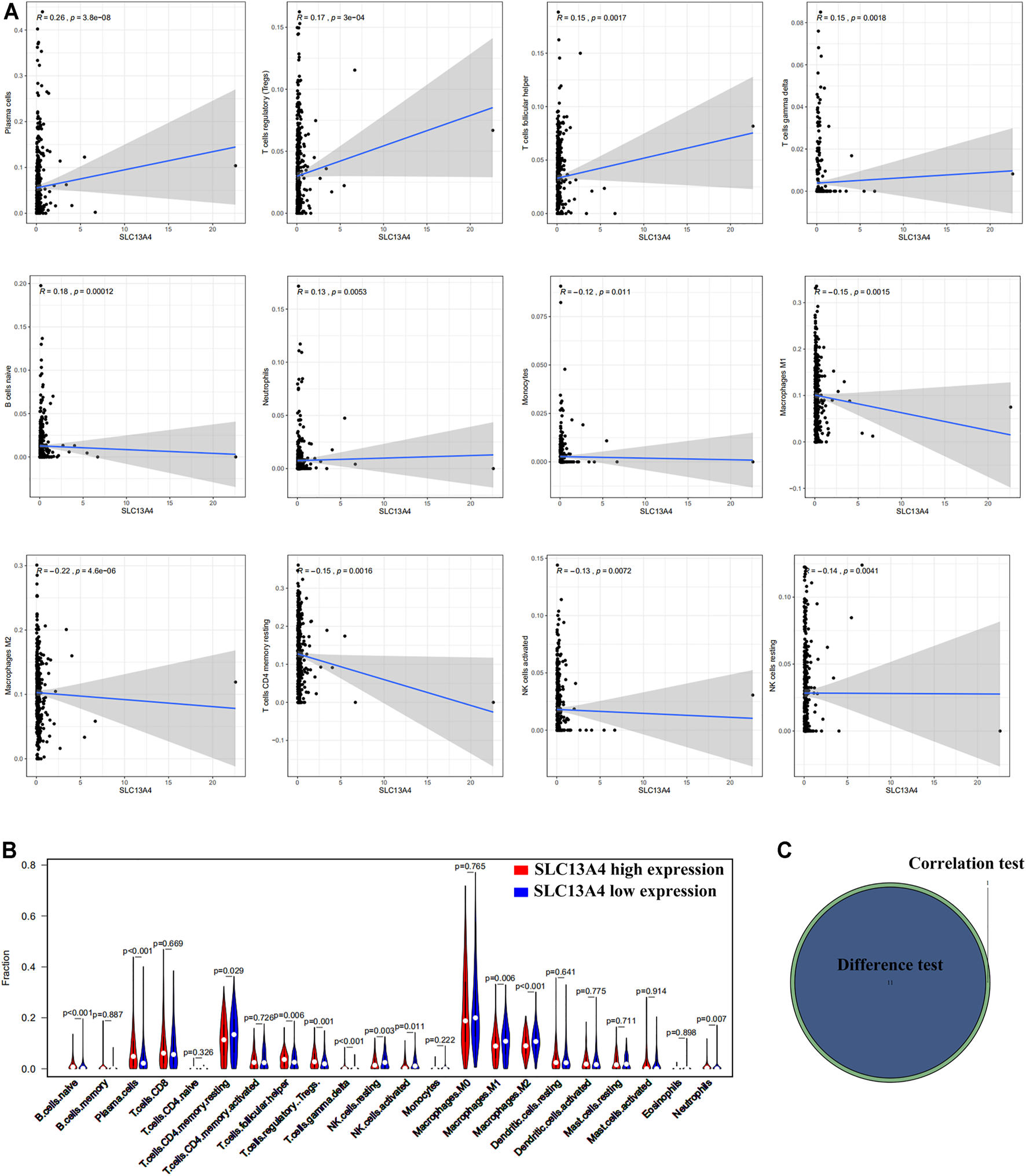
Correlation between TICs proportion and SLC13A4 expression. (A) Scatter plot showed the correlation of 12 kinds of TICs proportion with the SLC13A4 expression (p < 0.05). The gray line in each plot was fitted linear model indicating the proportion tropism of the immune cell along with SLC13A4 expression, and Pearson coefficient was used for the correlation test. (B) Violin plot showed the ratio differentiation of 21 kinds of immune cells in high and low SLC13A4 expression group in HNSC tumor sample, and Wilcoxon rank sum was used for the significance test. (C) The Venn diagram shows 11 TICs related to SLC13A4 expression.
Correlation Between SLC13A4 Expression and Modulation of Immunity and Tumor Pathways
In order to further understand the potential mechanism of SLC13A4 affecting the prognosis of HNSCC, patients were divided into a high- and a low-expression groups according to the median expression level of SLC13A4 for GSEA enrichment analysis. The results are shown in Figure 6. The genes in low SLC13A4 expression group were principally involved in immunity-related activities, viral diseases, typical tumor pathways and metabolism, including antigen processing and presentation, regulatory T cells, interleukin-family members, viral myocarditis, epithelial mesenchymal transition (EMT), glycosaminoglycan biosynthesis chondroitin sulfate, among others. In the high SLC13A4 expression group, mainly the metabolism-related pathways were enriched, including metabolisms of linoleic acid, retinol, arachidonic acid and alpha linolenic acid. However, there were no enrichment of genome sets involving immune-related activities and typical tumor pathways in the high SLC13A4 expression group, which further suggested that low SLC13A4 expression might be implicated in the process of cancer development and immune activities.
FIGURE 6
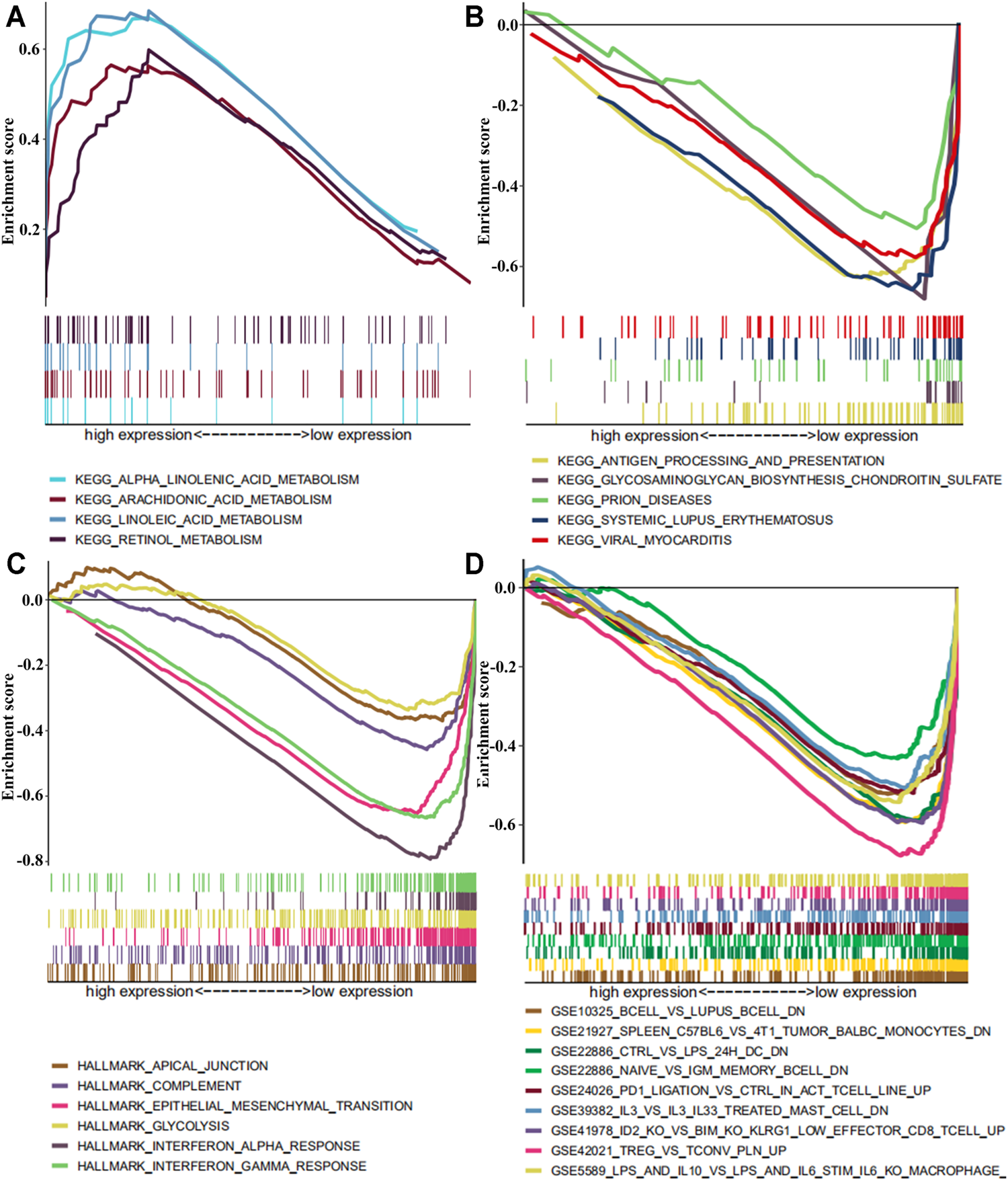
The GSEA enrichment analysis by expression level of SLC13A4 in HNSCC patients. Each row represents a specific set of genes with a unique color. (A) Enriched gene sets with high expression of SLC13A4 in KEGG. (B) Enriched gene sets with low expression of SLC13A4 in KEGG. (C) Enriched gene sets with low expression of SLC13A4 in HALLMARK. (D) Enriched gene sets with low expression of SLC13A4 in IMMUNE SIGNITURE and only a few major gene sets were shown (p < 0.01 and FDR ≤ 25%).
Discussion
SLC13A4 is a sulfate transporter and is also responsible for the anion transport of selenium and chromium [29]. At present, most studies focused on its roles in severe fetal abnormalities [11], nerve system development [15], and so on. However, sulfate metabolism disorders can cause a variety of diseases and might serve as a predictor for tumor diagnosis [18]. Squamous cell carcinoma is a cancer that arises from particular cells known as squamous cells. Squamous cells are found in the outer layer of skin and in the mucous membranes, which are the moist tissues that form the lining layer of body cavities, such as the airways and intestines. HNSCC is the most common tumor of the head and neck regions, developing in the mucous membrane of the mouth, nose and throat. This study showed that the expression of SLC13A4 was intimately related to the clinical features and prognosis of HNSCC. In HNSCC patients, the expression of SLC13A4 decreased with the increase of tumor pathological grade and clinical stage. The expression level of SLC13A4 in HPV-positive patients was higher than in HPV-negative patients, and the finding was consistent with the phenomenon that the survival rate of HPV-positive patients was higher than that of HPV-negative patients. In this study, only one patient was in M1 stage, so no further analysis was made. Kaplan Meier analysis exhibited that the high expressing SLC13A4 group had longer overall survival time, progression free survival time and disease specific survival time as compared with the low SLC13A4 expression group. On the basis of the aforementioned findings, we are led to believe that that SLC13A4 expression can serve as a predictor of tumor prognosis.
The infiltration of immune cells is one of the characteristic host immune responses to tumor cells and is strongly associated with the development and progression of cancer. Our results also indicated that SLC13A4 expression was significantly correlated with the infiltration of immune cells into HNSCC. SLC13A4 expression was negatively correlated with monocytes, M1 macrophages, M2 macrophages, resting CD4+ memory T cells, resting NK cells and activated NK cells, but was positively correlated with neutrophils, plasma cells, T follicular helper cells, gamma delta T cells, regulatory T cells and naive B cells. These results further confirmed that SLC13A4 level affected the immune activity of TME. SLC13A4 was closely related to the counts of macrophages, M1 and M2, which suggests that the expression of SLC13A4 might be a regulator of macrophage activation and polarization. SLC13A4 was positively correlated with the expression of Tregs, TFH and γδT cells, suggesting that SLC13A4 might regulate the T cell function of HNSCC. Our analysis also showed that SLC13A4 was associated with the degree of immune infiltration and the number of immune cells in HNSCC. It has been reported that the low ratio of M0 macrophages and high expression of Tregs contribute to the favorable prognosis of OS and DFS in HNSCC patients, which is consistent with our conclusion [30]. These findings suggest that SLC13A4 plays an important role in lymphocyte recruitment and control of immune infiltration in HNSCC.
Our GSEA enrichment analysis demonstrated that the genes of in low SLC13A4 expression group were mainly enriched in immunity, viral diseases, typical tumor pathways and metabolism, while genes in high SLC13A4 high expression group were mainly enriched in metabolism-related activities, such as metabolisms of linoleic acid, retinol, arachidonic acid and alpha linolenic acid. However, there was no genomic enrichment involving immunity and typical tumor pathways, suggesting that SLC13A4 might act as an indicator of TME transformation from metabolic dominance to immune dominance. The change of infiltrating immune cell components might be triggered by the growth of tumor tissue, and also affect the development of tumors, such as the tumor immune phenotype could predict their prognosis in patients with squamous cell carcinoma of the oral tongue and with squamous cell carcinoma of the head and neck [31,32]. The change of tumor immune microenvironment provides a new target for studying the development and progression of HNSCC. Immunotherapies aim to enhance the activity of the immune system, thereby eradicating cancer cells. Targeting immune checkpoints is a strategy extensively used in cancer therapy. The expression of SLC13A4 in HNSCC is negatively correlated with T cell exhaustion markers, including LAG3, TIM-3 and CTLA-4. Immune checkpoint inhibitors can block these T cell exhaustion markers signals and have proven their good anti-tumor effects, reverse depletion of T cells, and restore tumor microenvironment and tumor-infiltrating T cell functions [33–35]. These suggest that SLC13A4 might reverse T cell immunosuppression and inhibit tumor immune escape.
In conclusion, this study demonstrated that SLC13A4 might serve as a biomarker of HNSCC. Our study helps clarify the role of SLC13A4 in the prognosis of HNSCC patients and the transformation from metabolic dominance to immune one in TME, and pave the way to future application of the biomarker in the diagnosis and prognostic prediction of HNSCC [28].
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
M-LY: conceptualization, methodology, software, data curation, visualization, formal analysis, writing-original draft. J-HZ: conceptualization, software, visualization, validation, writing-review and editing. SL: validation, formal analysis. RZ: resources, investigation, data curation, writing-original draft. LW: supervision, funding acquisition, project administration, writing-review and editing.
Funding
This work was supported by grants from the Natural Sciences Foundation of Hubei Province, China (No: 2019CFB470), the Research Funds from the Health Commission of Hubei Province, China (No: WJ2019M168), the Fundamental Research Funds for the Central Universities of China (No: 2017KFYXJJ239) and the Key Laboratory Open Project Fund of Hubei Province (No: 2020swbx014). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgments
We acknowledge the TCGA, GEO ONCOMINE, TIMER, CIBERSORT databases for free use.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.por-journal.com/articles/10.3389/pore.2021.1609967/full#supplementary-material
Supplementary Figure S1Prognostic significance of SLC13A4 in HNSCC from GEO validation cohorts. Kaplan-Meier survival curves of overall survival in GSE41613 (A), recurrence-free survival in GSE2837 (B), disease specific survival in GSE27020 (C).
References
1.
Ferlay J Soerjomataram I Dikshit R Eser S Mathers C Rebelo M et al Cancer Incidence and Mortality Worldwide: Sources, Methods and Major Patterns in GLOBOCAN 2012. Int J Cancer (2015) 136(5):E359–E386. 10.1002/ijc.29210
2.
Miller KD Fidler-Benaoudia M Keegan TH Hipp HS Jemal A Siegel RL . Cancer Statistics for Adolescents and Young Adults, 2020. CA Cancer J Clin (2020) 70(1):443–59. 10.3322/caac.21637
3.
Solomon B Young RJ Rischin D . Head and Neck Squamous Cell Carcinoma: Genomics and Emerging Biomarkers for Immunomodulatory Cancer Treatments. Semin Cancer Biol (2018) 52(Pt 2):228–40. 10.1016/j.semcancer.2018.01.008
4.
Peltanova B Raudenska M Masarik M . Effect of Tumor Microenvironment on Pathogenesis of the Head and Neck Squamous Cell Carcinoma: a Systematic Review. Mol Cancer (2019) 18(1):63. 10.1186/s12943-019-0983-5
5.
Cohen EEW Soulières D Le Tourneau C Dinis J Licitra L Ahn MJ et al KEYNOTE-040 Investigators. Pembrolizumab versus Methotrexate, Docetaxel, or Cetuximab for Recurrent or Metastatic Head-And-Neck Squamous Cell Carcinoma (KEYNOTE-040): a Randomised, Open-Label, Phase 3 Study. Lancet (2019) 393(10167):156–67. 10.1016/S0140-6736(18)31999-8
6.
Ferris RL Blumenschein G Jr Fayette J Guigay J Colevas AD Licitra L et al Nivolumab vs Investigator's Choice in Recurrent or Metastatic Squamous Cell Carcinoma of the Head and Neck: 2-year Long-Term Survival Update of CheckMate 141 with Analyses by Tumor PD-L1 Expression. Oral Oncol (2018) 81:45–51. 10.1016/j.oraloncology.2018.04.008
7.
Bergeron MJ Clémençon B Hediger MA Markovich D . SLC13 Family of Na⁺-Coupled Di- and Tri-carboxylate/sulfate Transporters. Mol Aspects Med (2013) 34(2-3):299–312. 10.1016/j.mam.2012.12.001
8.
Marutani E Yamada M Ida T Tokuda K Ikeda K Kai S et al Thiosulfate Mediates Cytoprotective Effects of Hydrogen Sulfide against Neuronal Ischemia. J Am Heart Assoc (2015) 4(11):e002125. 10.1161/JAHA.115.002125
9.
Dawson PA Pirlo KJ Steane SE Kunzelmann K Chien YJ Markovich D . Molecular Cloning and Characterization of the Mouse Na+ Sulfate Cotransporter Gene (Slc13a4): Structure and Expression. Genes Genet Syst (2006) 81(4):265–72. 10.1266/ggs.81.265
10.
Markovich D Regeer RR Kunzelmann K Dawson PA . Functional Characterization and Genomic Organization of the Human Na+-Sulfate Cotransporter hNaS2 Gene (SLC13A4). Biochem Biophysical Res Commun (2005) 326(4):729–34. 10.1016/j.bbrc.2004.11.102
11.
Rakoczy J Zhang Z Bowling FG Dawson PA Simmons DG . Loss of the Sulfate Transporter Slc13a4 in Placenta Causes Severe Fetal Abnormalities and Death in Mice. Cell Res (2015) 25(11):1273–6. 10.1038/cr.2015.100
12.
Zhang Z Aung ZT Simmons DG Dawson PA . Molecular Analysis of Sequence and Splice Variants of the Human SLC13A4 Sulfate Transporter. Mol Genet Metab (2017) 121(1):35–42. 10.1016/j.ymgme.2017.03.010
13.
Hästbacka J de la Chapelle A Mahtani MM Clines G Reeve-Daly MP Daly M et al The Diastrophic Dysplasia Gene Encodes a Novel Sulfate Transporter: Positional Cloning by fine-structure Linkage Disequilibrium Mapping. Cell (1994) 78(6):1073–87. 10.1016/0092-8674(94)90281-x
14.
Demydchuk M Hill CH Zhou A Bunkóczi G Stein PE Marchesan D et al Insights into Hunter Syndrome from the Structure of Iduronate-2-Sulfatase. Nat Commun (2017) 8:15786. 10.1038/ncomms15786
15.
Zhang Z Dawson PA Piper M Simmons DG . Postnatal N-Acetylcysteine Administration Rescues Impaired Social Behaviors and Neurogenesis in Slc13a4 Haploinsufficient Mice. EBioMedicine (2019) 43:435–46. 10.1016/j.ebiom.2019.03.081
16.
Markovich D . Physiological Roles and Regulation of Mammalian Sulfate Transporters. Physiol Rev (2001) 81(4):1499–533. 10.1152/physrev.2001.81.4.1499
17.
Dawson PA . Role of Sulphate in Development. Reproduction (2013) 146(3):R81–R89. 10.1530/rep-13-0056
18.
Chwatko G Forma E Wilkosz J Głowacki R Jóźwiak P Różański W et al Thiosulfate in Urine as a Facilitator in the Diagnosis of Prostate Cancer for Patients with Prostate-specific Antigen Less or Equal 10 ng/mL. Clin Chem Lab Med (2013) 51(9):1825–31. 10.1515/cclm-2013-0069
19.
Baskar R Bian J . Hydrogen Sulfide Gas Has Cell Growth Regulatory Role. Eur J Pharmacol (2011) 656(1-3):5–9. 10.1016/j.ejphar.2011.01.052
20.
Cai WJ Wang MJ Ju LH Wang C Zhu YC . Hydrogen Sulfide Induces Human colon Cancer Cell Proliferation: Role of Akt, ERK and P21. Cell. Biol. Int. (2010) 34(6):565–72. 10.1042/cbi20090368
21.
Cao Q Zhang L Yang G Xu C Wang R . Butyrate-stimulated H2S Production in colon Cancer Cells. Antioxid Redox Signaling (2010) 12(9):1101–9. 10.1089/ars.2009.2915
22.
Wang Z Jensen MA Zenklusen JC . A Practical Guide to the Cancer Genome Atlas (TCGA). Methods Mol Biol (2016) 1418:111–41. 10.1007/978-1-4939-3578-9_6
23.
Jiao Y Fu Z Li Y Meng L Liu Y . High EIF2B5 mRNA Expression and its Prognostic Significance in Liver Cancer: A Study Based on the TCGA and GEO Database. Cmar (2018) Vol. 10:6003–14. 10.2147/cmar.s185459
24.
Rhodes DR Kalyana-Sundaram S Mahavisno V Varambally R Yu J Briggs BB et al Oncomine 3.0: Genes, Pathways, and Networks in a Collection of 18,000 Cancer Gene Expression Profiles. Neoplasia (2007) 9(2):166–80. 10.1593/neo.07112
25.
Li T Fan J Wang B Traugh N Chen Q Liu JS et al TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res (2017) 77(21):e108–e110. 10.1158/0008-5472.can-17-0307
26.
Uhlén M Fagerberg L Hallström BM Lindskog C Oksvold P Mardinoglu A et al Tissue-based Map of the Human Proteome. Science (2015) 347(6220):1260419. 10.1126/science.1260419
27.
Newman AM Liu CL Green MR Gentles AJ Feng W Xu Y et al Robust Enumeration of Cell Subsets from Tissue Expression Profiles. Nat Methods (2015) 12(5):453–7. 10.1038/nmeth.3337
28.
Subramanian A Tamayo P Mootha VK Mukherjee S Ebert BL Gillette MA et al Gene Set Enrichment Analysis: a Knowledge-Based Approach for Interpreting Genome-wide Expression Profiles. Proc Natl Acad Sci (2005) 102(43):15545–50. 10.1073/pnas.0506580102
29.
Miyauchi S Srinivas SR Fei YJ Gopal E Umapathy NS Wang H et al Functional Characteristics of NaS2, a Placenta-specific Na+-Coupled Transporter for Sulfate and Oxyanions of the Micronutrients Selenium and Chromium. Placenta (2006) 27(6-7):550–9. 10.1016/j.placenta.2005.05.004
30.
Liang B Tao Y Wang T . Profiles of Immune Cell Infiltration in Head and Neck Squamous Carcinoma. Biosci Rep (2020) 40(2):BSR20192724. 10.1042/BSR20192724
31.
Hanna GJ Liu H Jones RE Bacay AF Lizotte PH Ivanova EV et al Defining an Inflamed Tumor Immunophenotype in Recurrent, Metastatic Squamous Cell Carcinoma of the Head and Neck. Oral Oncol (2017) 67:61–9. 10.1016/j.oraloncology.2017.02.005
32.
Troiano G Rubini C Togni L Caponio VCA Zhurakivska K Santarelli A et al The Immune Phenotype of Tongue Squamous Cell Carcinoma Predicts Early Relapse and Poor Prognosis. Cancer Med (2020) 9(22):8333–44. 10.1002/cam4.3440
33.
Schoenfeld JD Hanna GJ Jo VY Rawal B Chen Y-H Catalano PS et al Neoadjuvant Nivolumab or Nivolumab Plus Ipilimumab in Untreated Oral Cavity Squamous Cell Carcinoma. JAMA Oncol (2020) 6(10):1563–70. 10.1001/jamaoncol.2020.2955
34.
Jiang H Ni H Zhang P Guo X Wu M Shen H et al PD-L1/LAG-3 Bispecific Antibody Enhances Tumor-specific Immunity. Oncoimmunology (2021) 10(1):1943180. 10.1080/2162402x.2021.1943180
35.
Liu J-F Wu L Yang L-L Deng W-W Mao L Wu H et al Blockade of TIM3 Relieves Immunosuppression through Reducing Regulatory T Cells in Head and Neck Cancer. J Exp Clin Cancer Res (2018) 37(1):44. 10.1186/s13046-018-0713-7
Summary
Keywords
biomarker, tumor-infiltrating immune cells, SLC13A4, head and neck squamous cell carcinoma, database analysis
Citation
Yang M-L, Zhang J-H, Li S, Zhu R and Wang L (2021) SLC13A4 Might Serve as a Prognostic Biomarker and be Correlated with Immune Infiltration into Head and Neck Squamous Cell Carcinoma. Pathol. Oncol. Res. 27:1609967. doi: 10.3389/pore.2021.1609967
Received
18 July 2021
Accepted
05 October 2021
Published
10 November 2021
Volume
27 - 2021
Edited by
Andrea Ladányi, National Institute of Oncology (NIO), Hungary
Updates
Copyright
© 2021 Yang, Zhang, Li, Zhu and Wang.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rui Zhu, zhur@hust.edu.cn; Li Wang, wangli1229@hust.edu.cn
†These authors have contributed equally to this work
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.